Graph-Based Retriever Captures the Long Tail of Biomedical Knowledge
Paper and Code
Feb 19, 2024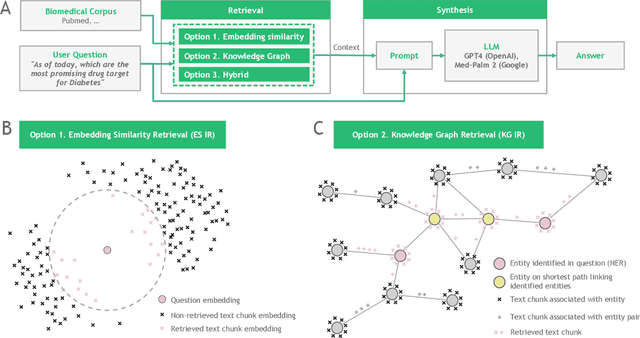
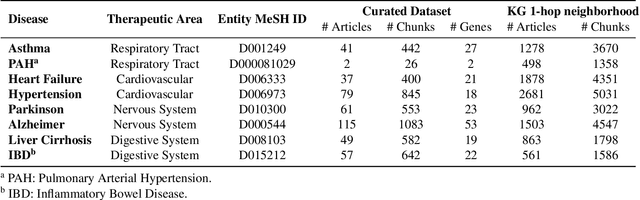
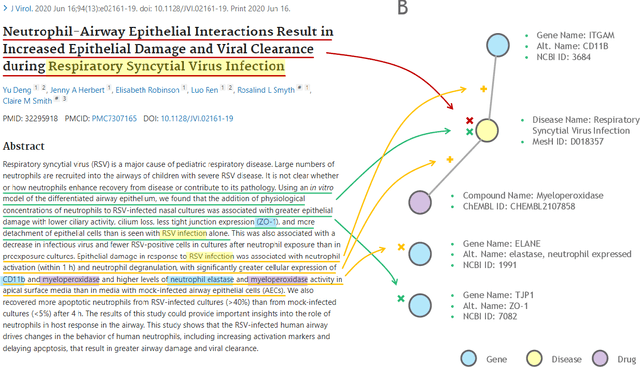
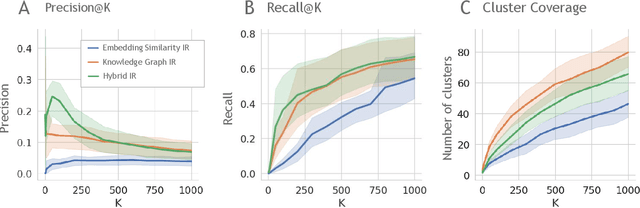
Large language models (LLMs) are transforming the way information is retrieved with vast amounts of knowledge being summarized and presented via natural language conversations. Yet, LLMs are prone to highlight the most frequently seen pieces of information from the training set and to neglect the rare ones. In the field of biomedical research, latest discoveries are key to academic and industrial actors and are obscured by the abundance of an ever-increasing literature corpus (the information overload problem). Surfacing new associations between biomedical entities, e.g., drugs, genes, diseases, with LLMs becomes a challenge of capturing the long-tail knowledge of the biomedical scientific production. To overcome this challenge, Retrieval Augmented Generation (RAG) has been proposed to alleviate some of the shortcomings of LLMs by augmenting the prompts with context retrieved from external datasets. RAG methods typically select the context via maximum similarity search over text embeddings. In this study, we show that RAG methods leave out a significant proportion of relevant information due to clusters of over-represented concepts in the biomedical literature. We introduce a novel information-retrieval method that leverages a knowledge graph to downsample these clusters and mitigate the information overload problem. Its retrieval performance is about twice better than embedding similarity alternatives on both precision and recall. Finally, we demonstrate that both embedding similarity and knowledge graph retrieval methods can be advantageously combined into a hybrid model that outperforms both, enabling potential improvements to biomedical question-answering models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge