Genie: A new, fast, and outlier-resistant hierarchical clustering algorithm
Paper and Code
Sep 13, 2022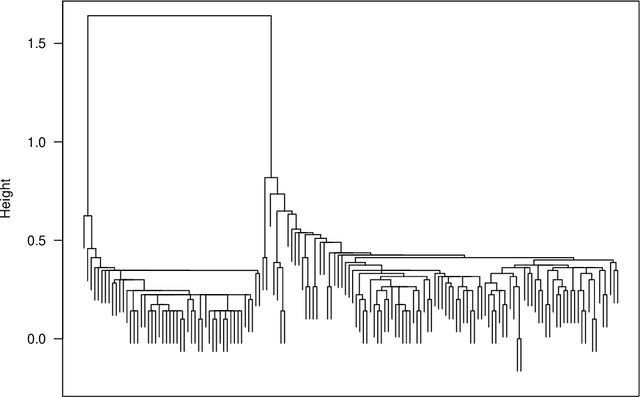
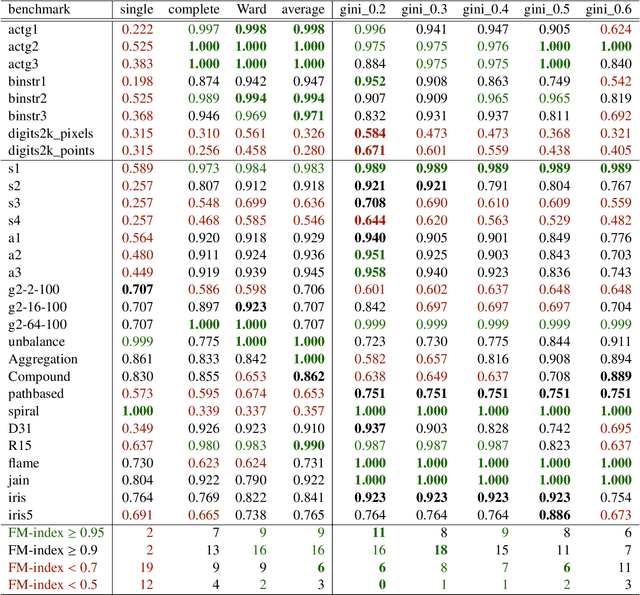
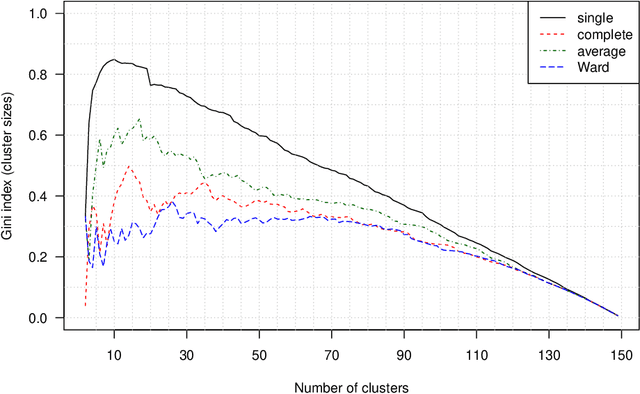
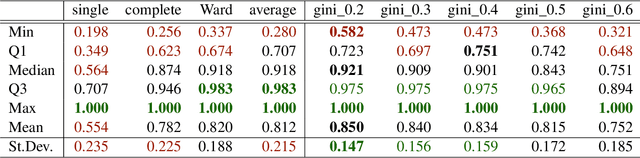
The time needed to apply a hierarchical clustering algorithm is most often dominated by the number of computations of a pairwise dissimilarity measure. Such a constraint, for larger data sets, puts at a disadvantage the use of all the classical linkage criteria but the single linkage one. However, it is known that the single linkage clustering algorithm is very sensitive to outliers, produces highly skewed dendrograms, and therefore usually does not reflect the true underlying data structure -- unless the clusters are well-separated. To overcome its limitations, we propose a new hierarchical clustering linkage criterion called Genie. Namely, our algorithm links two clusters in such a way that a chosen economic inequity measure (e.g., the Gini- or Bonferroni-index) of the cluster sizes does not drastically increase above a given threshold. The presented benchmarks indicate a high practical usefulness of the introduced method: it most often outperforms the Ward or average linkage in terms of the clustering quality while retaining the single linkage's speed. The Genie algorithm is easily parallelizable and thus may be run on multiple threads to speed up its execution even further. Its memory overhead is small: there is no need to precompute the complete distance matrix to perform the computations in order to obtain a desired clustering. It can be applied on arbitrary spaces equipped with a dissimilarity measure, e.g., on real vectors, DNA or protein sequences, images, rankings, informetric data, etc. A reference implementation of the algorithm has been included in the open source 'genie' package for R. See also https://genieclust.gagolewski.com for a new implementation (genieclust) -- available for both R and Python.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge