Generating Knowledge Graphs by Employing Natural Language Processing and Machine Learning Techniques within the Scholarly Domain
Paper and Code
Oct 28, 2020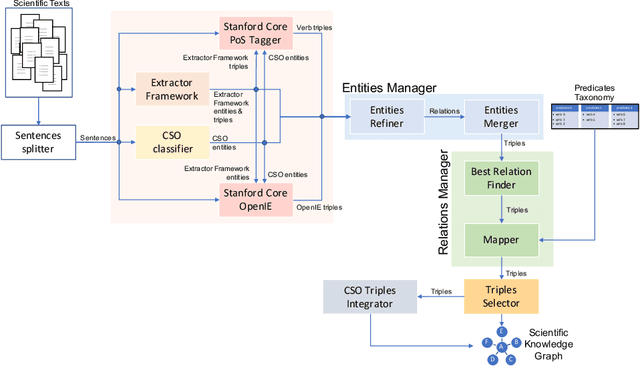
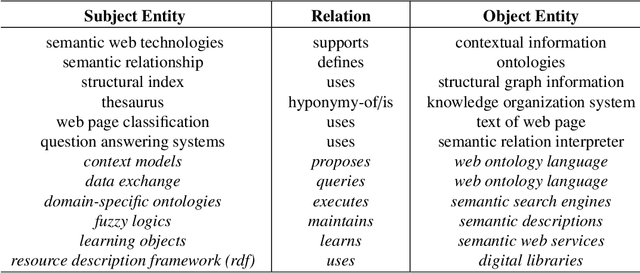
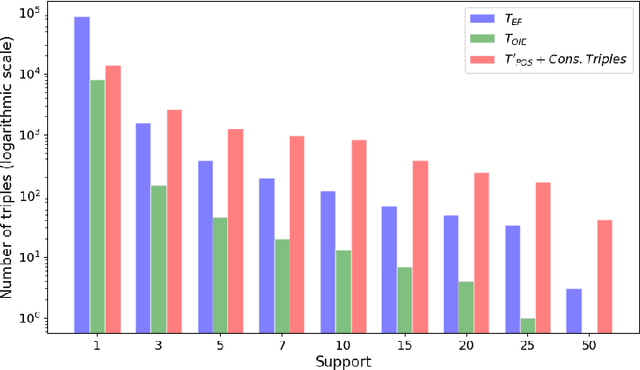
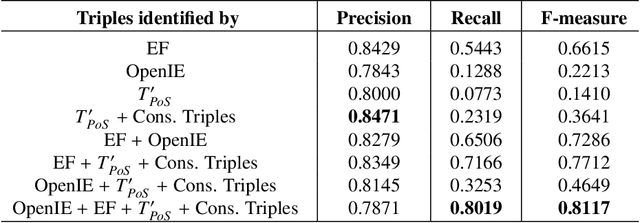
The continuous growth of scientific literature brings innovations and, at the same time, raises new challenges. One of them is related to the fact that its analysis has become difficult due to the high volume of published papers for which manual effort for annotations and management is required. Novel technological infrastructures are needed to help researchers, research policy makers, and companies to time-efficiently browse, analyse, and forecast scientific research. Knowledge graphs i.e., large networks of entities and relationships, have proved to be effective solution in this space. Scientific knowledge graphs focus on the scholarly domain and typically contain metadata describing research publications such as authors, venues, organizations, research topics, and citations. However, the current generation of knowledge graphs lacks of an explicit representation of the knowledge presented in the research papers. As such, in this paper, we present a new architecture that takes advantage of Natural Language Processing and Machine Learning methods for extracting entities and relationships from research publications and integrates them in a large-scale knowledge graph. Within this research work, we i) tackle the challenge of knowledge extraction by employing several state-of-the-art Natural Language Processing and Text Mining tools, ii) describe an approach for integrating entities and relationships generated by these tools, iii) show the advantage of such an hybrid system over alternative approaches, and vi) as a chosen use case, we generated a scientific knowledge graph including 109,105 triples, extracted from 26,827 abstracts of papers within the Semantic Web domain. As our approach is general and can be applied to any domain, we expect that it can facilitate the management, analysis, dissemination, and processing of scientific knowledge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge