Generalized Bayesian Inference for Scientific Simulators via Amortized Cost Estimation
Paper and Code
May 24, 2023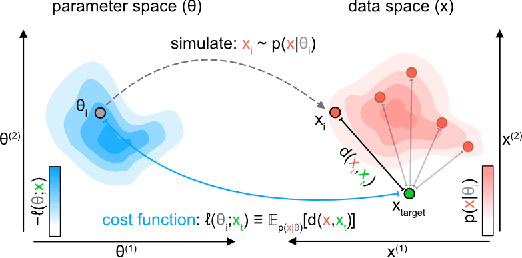
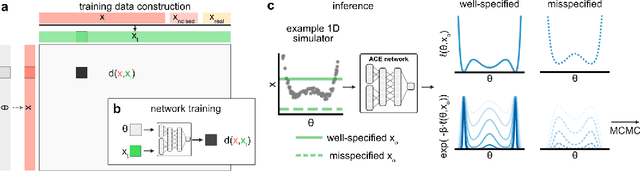
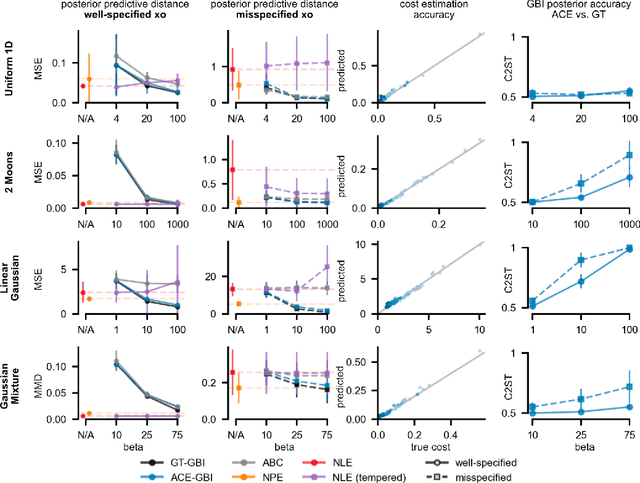
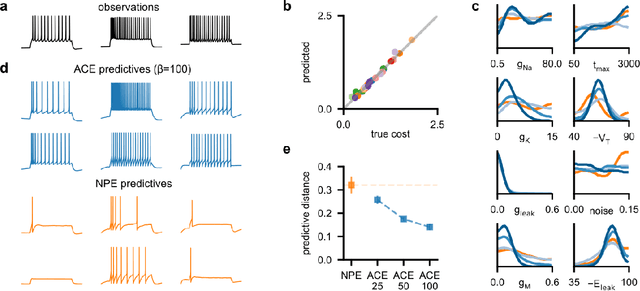
Simulation-based inference (SBI) enables amortized Bayesian inference for simulators with implicit likelihoods. But when we are primarily interested in the quality of predictive simulations, or when the model cannot exactly reproduce the observed data (i.e., is misspecified), targeting the Bayesian posterior may be overly restrictive. Generalized Bayesian Inference (GBI) aims to robustify inference for (misspecified) simulator models, replacing the likelihood-function with a cost function that evaluates the goodness of parameters relative to data. However, GBI methods generally require running multiple simulations to estimate the cost function at each parameter value during inference, making the approach computationally infeasible for even moderately complex simulators. Here, we propose amortized cost estimation (ACE) for GBI to address this challenge: We train a neural network to approximate the cost function, which we define as the expected distance between simulations produced by a parameter and observed data. The trained network can then be used with MCMC to infer GBI posteriors for any observation without running additional simulations. We show that, on several benchmark tasks, ACE accurately predicts cost and provides predictive simulations that are closer to synthetic observations than other SBI methods, especially for misspecified simulators. Finally, we apply ACE to infer parameters of the Hodgkin-Huxley model given real intracellular recordings from the Allen Cell Types Database. ACE identifies better data-matching parameters while being an order of magnitude more simulation-efficient than a standard SBI method. In summary, ACE combines the strengths of SBI methods and GBI to perform robust and simulation-amortized inference for scientific simulators.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge