Gene selection with guided regularized random forest
Paper and Code
Jun 20, 2013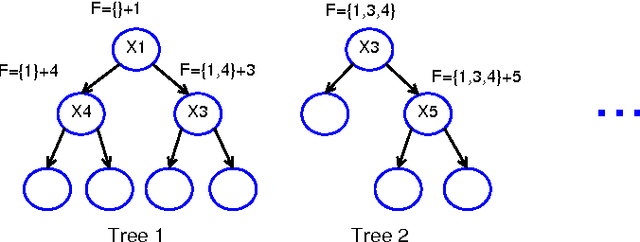

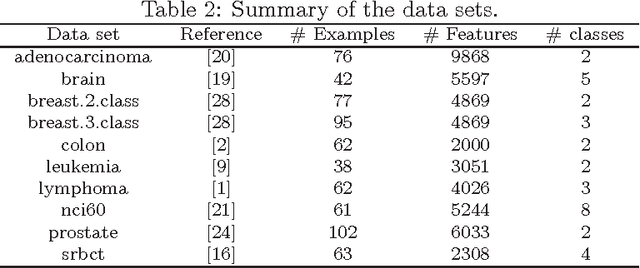
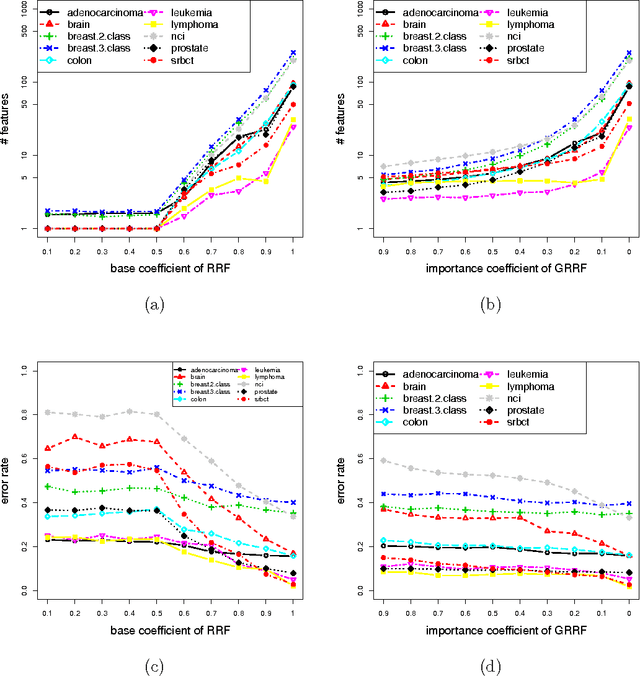
The regularized random forest (RRF) was recently proposed for feature selection by building only one ensemble. In RRF the features are evaluated on a part of the training data at each tree node. We derive an upper bound for the number of distinct Gini information gain values in a node, and show that many features can share the same information gain at a node with a small number of instances and a large number of features. Therefore, in a node with a small number of instances, RRF is likely to select a feature not strongly relevant. Here an enhanced RRF, referred to as the guided RRF (GRRF), is proposed. In GRRF, the importance scores from an ordinary random forest (RF) are used to guide the feature selection process in RRF. Experiments on 10 gene data sets show that the accuracy performance of GRRF is, in general, more robust than RRF when their parameters change. GRRF is computationally efficient, can select compact feature subsets, and has competitive accuracy performance, compared to RRF, varSelRF and LASSO logistic regression (with evaluations from an RF classifier). Also, RF applied to the features selected by RRF with the minimal regularization outperforms RF applied to all the features for most of the data sets considered here. Therefore, if accuracy is considered more important than the size of the feature subset, RRF with the minimal regularization may be considered. We use the accuracy performance of RF, a strong classifier, to evaluate feature selection methods, and illustrate that weak classifiers are less capable of capturing the information contained in a feature subset. Both RRF and GRRF were implemented in the "RRF" R package available at CRAN, the official R package archive.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge