FsNet: Feature Selection Network on High-dimensional Biological Data
Paper and Code
Jan 23, 2020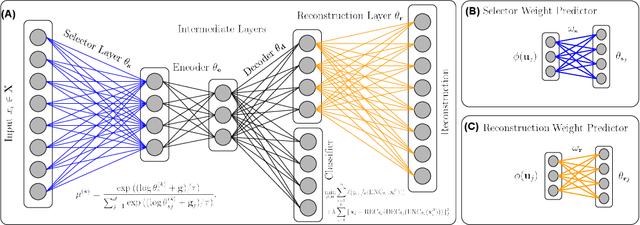
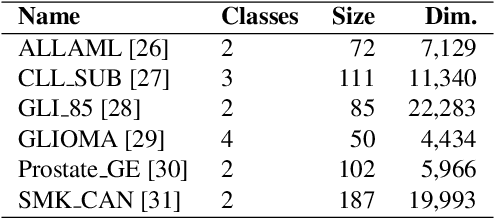
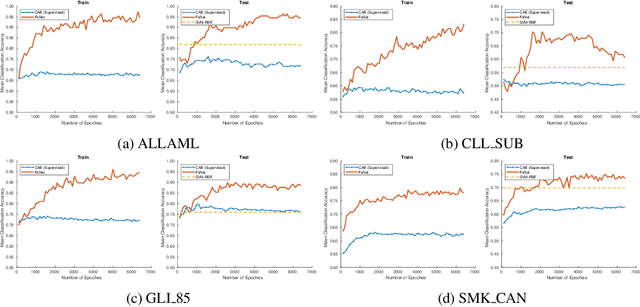
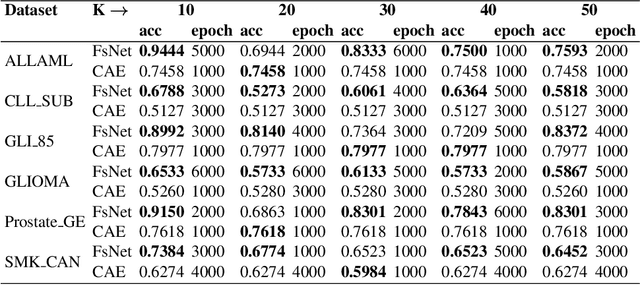
Biological data are generally high-dimensional and require efficient machine learning methods that are well generalized and scalable to discover their complex nonlinear patterns. The recent advances in the domain of artificial intelligence and machine learning can be attributed to deep neural networks (DNNs) because they accomplish a variety of tasks in computer vision and natural language processing. However, standard DNNs are not suitable for handling high-dimensional data and data with small number of samples because they require a large pool of computing resources as well as plenty of samples to learn a large number of parameters. In particular, although interpretability is important for high-dimensional biological data such as gene expression data, a nonlinear feature selection algorithm for DNN models has not been fully investigated. In this paper, we propose a novel nonlinear feature selection method called the Feature Selection Network (FsNet), which is a scalable concrete neural network architecture, under high-dimensional and small number of samples setups. Specifically, our network consists of a selector layer that uses a concrete random variable for discrete feature selection and a supervised deep neural network regularized with the reconstruction loss. Because a large number of parameters in the selector and reconstruction layer can easily cause overfitting under a limited number of samples, we use two tiny networks to predict the large virtual weight matrices of the selector and reconstruction layers. The experimental results on several real-world high-dimensional biological datasets demonstrate the efficacy of the proposed approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge