FLOWR: Flow Matching for Structure-Aware De Novo, Interaction- and Fragment-Based Ligand Generation
Paper and Code
Apr 14, 2025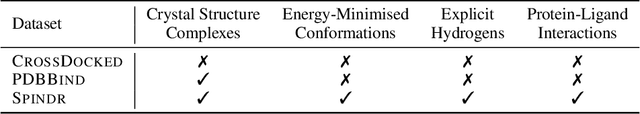
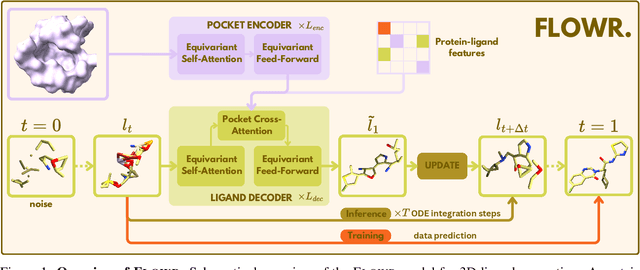
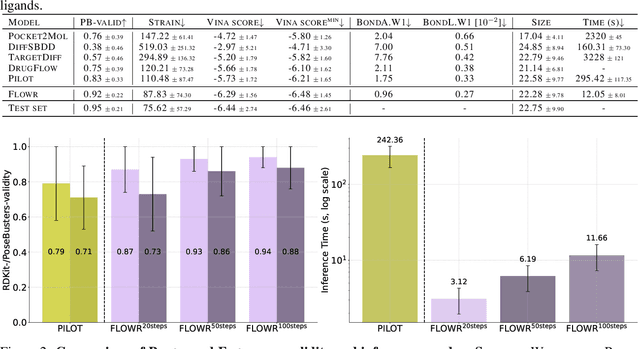
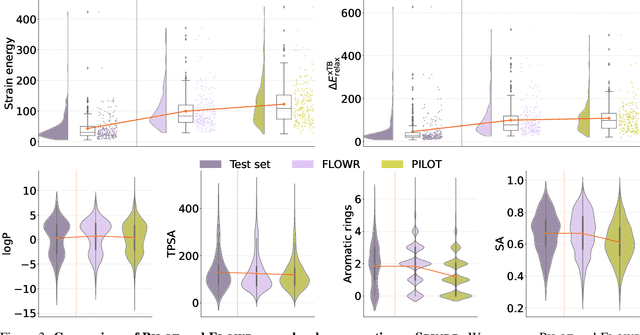
We introduce FLOWR, a novel structure-based framework for the generation and optimization of three-dimensional ligands. FLOWR integrates continuous and categorical flow matching with equivariant optimal transport, enhanced by an efficient protein pocket conditioning. Alongside FLOWR, we present SPINDR, a thoroughly curated dataset comprising ligand-pocket co-crystal complexes specifically designed to address existing data quality issues. Empirical evaluations demonstrate that FLOWR surpasses current state-of-the-art diffusion- and flow-based methods in terms of PoseBusters-validity, pose accuracy, and interaction recovery, while offering a significant inference speedup, achieving up to 70-fold faster performance. In addition, we introduce FLOWR.multi, a highly accurate multi-purpose model allowing for the targeted sampling of novel ligands that adhere to predefined interaction profiles and chemical substructures for fragment-based design without the need of re-training or any re-sampling strategies
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge