Exploiting Ligand Additivity for Transferable Machine Learning of Multireference Character Across Known Transition Metal Complex Ligands
Paper and Code
May 05, 2022
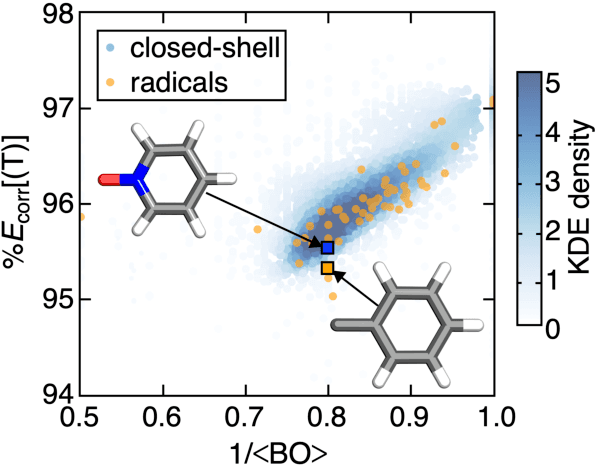

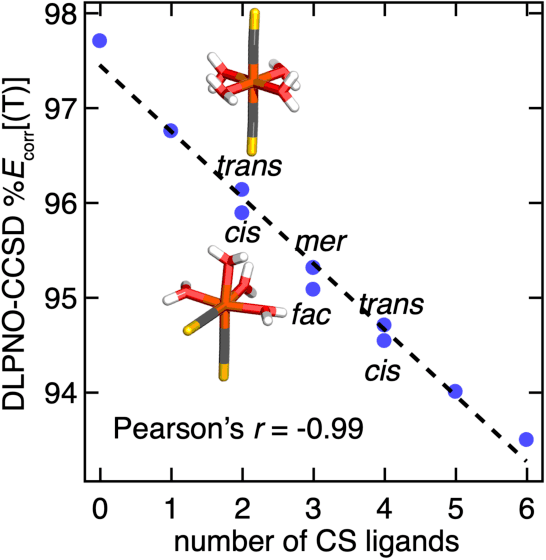
Accurate virtual high-throughput screening (VHTS) of transition metal complexes (TMCs) remains challenging due to the possibility of high multi-reference (MR) character that complicates property evaluation. We compute MR diagnostics for over 5,000 ligands present in previously synthesized transition metal complexes in the Cambridge Structural Database (CSD). To accomplish this task, we introduce an iterative approach for consistent ligand charge assignment for ligands in the CSD. Across this set, we observe that MR character correlates linearly with the inverse value of the averaged bond order over all bonds in the molecule. We then demonstrate that ligand additivity of MR character holds in TMCs, which suggests that the TMC MR character can be inferred from the sum of the MR character of the ligands. Encouraged by this observation, we leverage ligand additivity and develop a ligand-derived machine learning representation to train neural networks to predict the MR character of TMCs from properties of the constituent ligands. This approach yields models with excellent performance and superior transferability to unseen ligand chemistry and compositions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge