Exemplar Auditing for Multi-Label Biomedical Text Classification
Paper and Code
Apr 07, 2020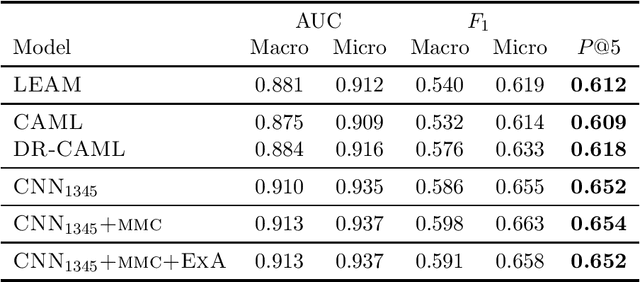
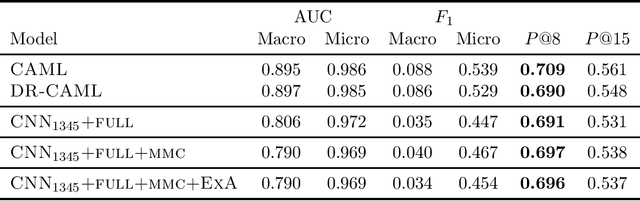
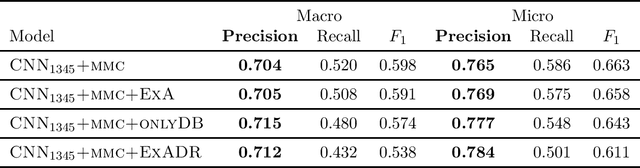
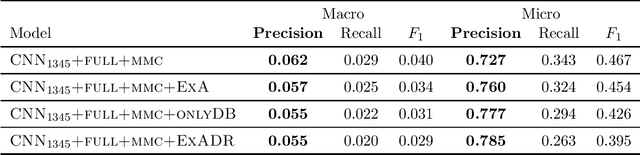
Many practical applications of AI in medicine consist of semi-supervised discovery: The investigator aims to identify features of interest at a resolution more fine-grained than that of the available human labels. This is often the scenario faced in healthcare applications as coarse, high-level labels (e.g., billing codes) are often the only sources that are readily available. These challenges are compounded for modalities such as text, where the feature space is very high-dimensional, and often contains considerable amounts of noise. In this work, we generalize a recently proposed zero-shot sequence labeling method, "binary labeling via a convolutional decomposition", to the case where the available document-level human labels are themselves relatively high-dimensional. The approach yields classification with "introspection", relating the fine-grained features of an inference-time prediction to their nearest neighbors from the training set, under the model. The approach is effective, yet parsimonious, as demonstrated on a well-studied MIMIC-III multi-label classification task of electronic health record data, and is useful as a tool for organizing the analysis of neural model predictions and high-dimensional datasets. Our proposed approach yields both a competitively effective classification model and an interrogation mechanism to aid healthcare workers in understanding the salient features that drive the model's predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge