EvoVGM: A Deep Variational Generative Model for Evolutionary Parameter Estimation
Paper and Code
May 25, 2022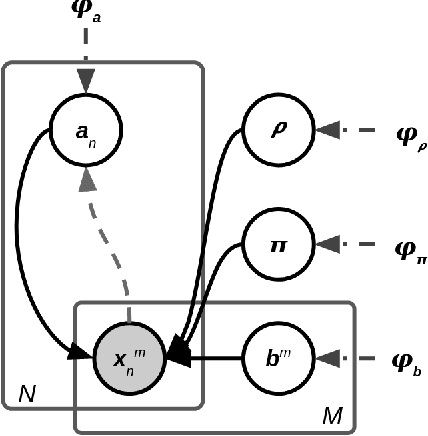
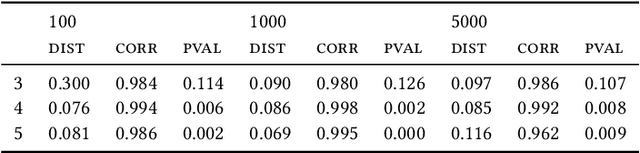
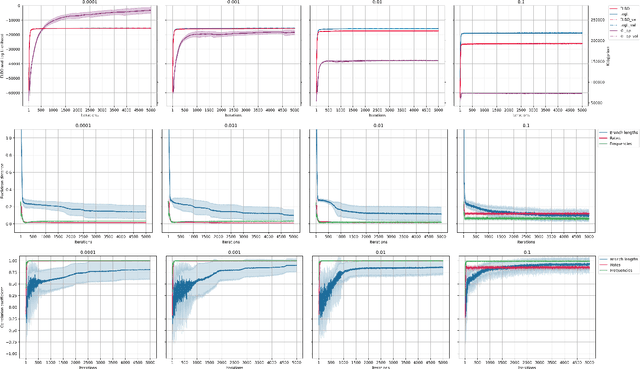
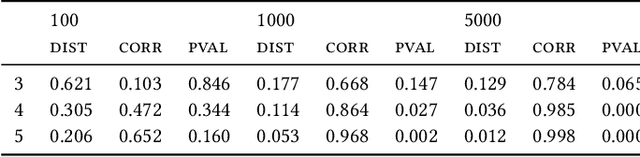
Most evolutionary-oriented deep generative models do not explicitly consider the underlying evolutionary dynamics of biological sequences as it is performed within the Bayesian phylogenetic inference framework. In this study, we propose a method for a deep variational Bayesian generative model that jointly approximates the true posterior of local biological evolutionary parameters and generates sequence alignments. Moreover, it is instantiated and tuned for continuous-time Markov chain substitution models such as JC69 and GTR. We train the model via a low-variance variational objective function and a gradient ascent algorithm. Here, we show the consistency and effectiveness of the method on synthetic sequence alignments simulated with several evolutionary scenarios and on a real virus sequence alignment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge