Equivariant Graph Neural Networks for 3D Macromolecular Structure
Paper and Code
Jun 07, 2021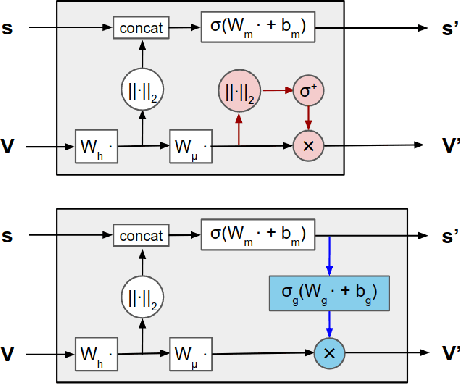
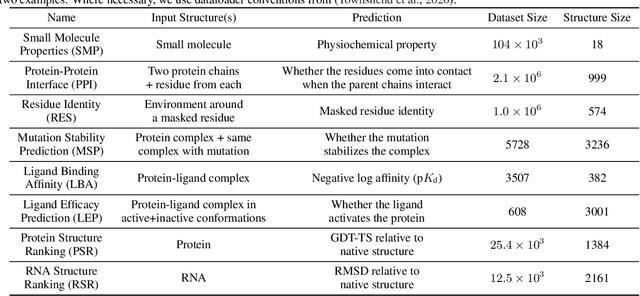

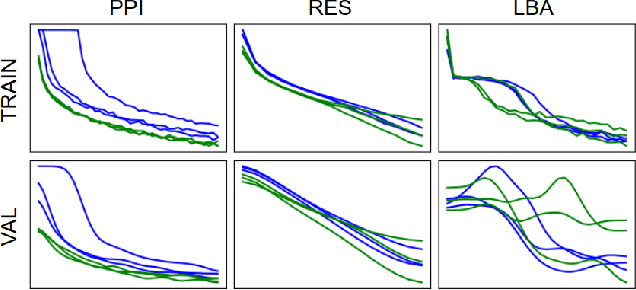
Representing and reasoning about 3D structures of macromolecules is emerging as a distinct challenge in machine learning. Here, we extend recent work on geometric vector perceptrons and apply equivariant graph neural networks to a wide range of tasks from structural biology. Our method outperforms all reference architectures on 4 out of 8 tasks in the ATOM3D benchmark and broadly improves over rotation-invariant graph neural networks. We also demonstrate that transfer learning can improve performance in learning from macromolecular structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge