End-to-End Reaction Field Energy Modeling via Deep Learning based Voxel-to-voxel Transform
Paper and Code
Oct 04, 2024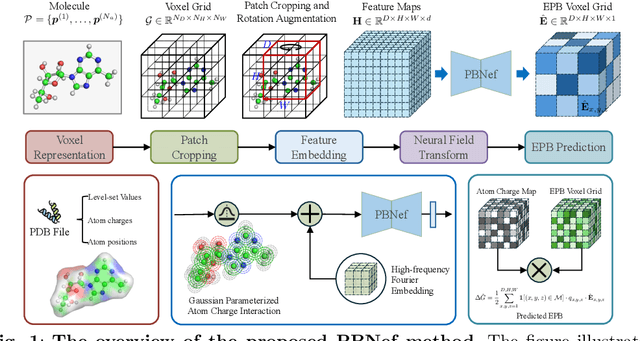
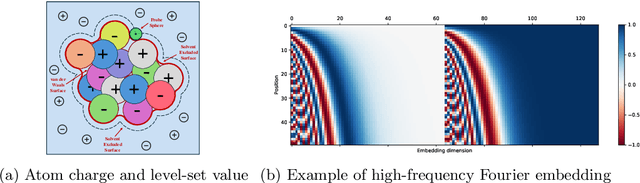
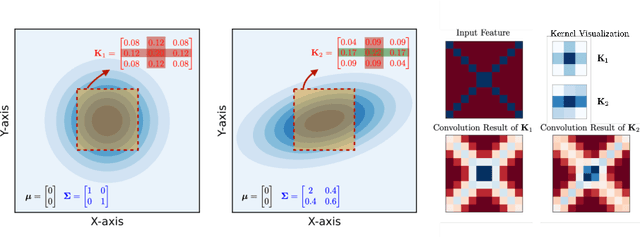
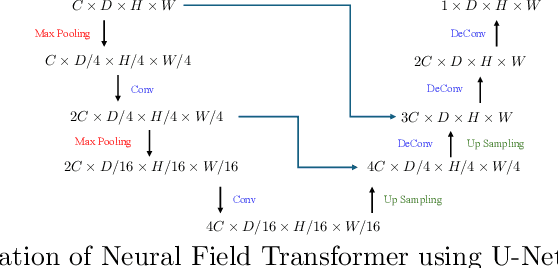
In computational biochemistry and biophysics, understanding the role of electrostatic interactions is crucial for elucidating the structure, dynamics, and function of biomolecules. The Poisson-Boltzmann (PB) equation is a foundational tool for modeling these interactions by describing the electrostatic potential in and around charged molecules. However, solving the PB equation presents significant computational challenges due to the complexity of biomolecular surfaces and the need to account for mobile ions. While traditional numerical methods for solving the PB equation are accurate, they are computationally expensive and scale poorly with increasing system size. To address these challenges, we introduce PBNeF, a novel machine learning approach inspired by recent advancements in neural network-based partial differential equation solvers. Our method formulates the input and boundary electrostatic conditions of the PB equation into a learnable voxel representation, enabling the use of a neural field transformer to predict the PB solution and, subsequently, the reaction field potential energy. Extensive experiments demonstrate that PBNeF achieves over a 100-fold speedup compared to traditional PB solvers, while maintaining accuracy comparable to the Generalized Born (GB) model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge