Embedded-physics machine learning for coarse-graining and collective variable discovery without data
Paper and Code
Feb 24, 2020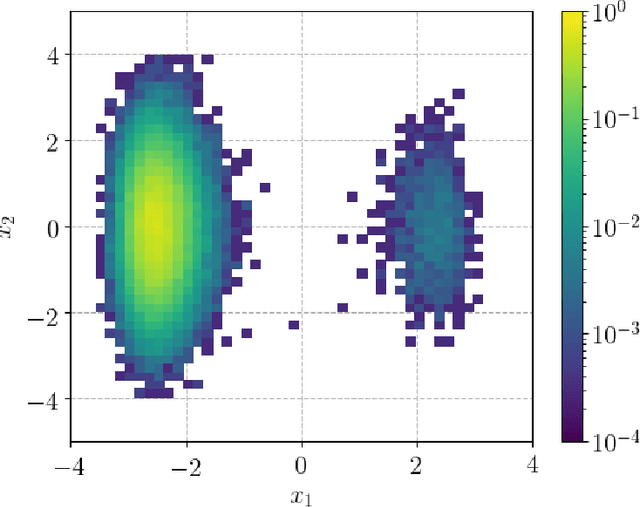
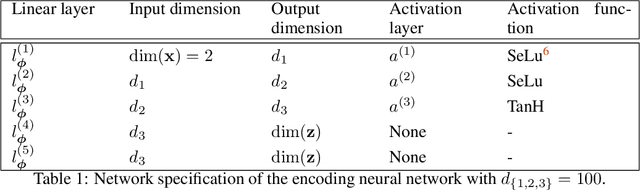
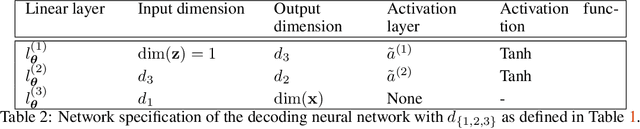

We present a novel learning framework that consistently embeds underlying physics while bypassing a significant drawback of most modern, data-driven coarse-grained approaches in the context of molecular dynamics (MD), i.e., the availability of big data. The generation of a sufficiently large training dataset poses a computationally demanding task, while complete coverage of the atomistic configuration space is not guaranteed. As a result, the explorative capabilities of data-driven coarse-grained models are limited and may yield biased "predictive" tools. We propose a novel objective based on reverse Kullback-Leibler divergence that fully incorporates the available physics in the form of the atomistic force field. Rather than separating model learning from the data-generation procedure - the latter relies on simulating atomistic motions governed by force fields - we query the atomistic force field at sample configurations proposed by the predictive coarse-grained model. Thus, learning relies on the evaluation of the force field but does not require any MD simulation. The resulting generative coarse-grained model serves as an efficient surrogate model for predicting atomistic configurations and estimating relevant observables. Beyond obtaining a predictive coarse-grained model, we demonstrate that in the discovered lower-dimensional representation, the collective variables (CVs) are related to physicochemical properties, which are essential for gaining understanding of unexplored complex systems. We demonstrate the algorithmic advances in terms of predictive ability and the physical meaning of the revealed CVs for a bimodal potential energy function and the alanine dipeptide.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge