DeepSet SimCLR: Self-supervised deep sets for improved pathology representation learning
Paper and Code
Feb 23, 2024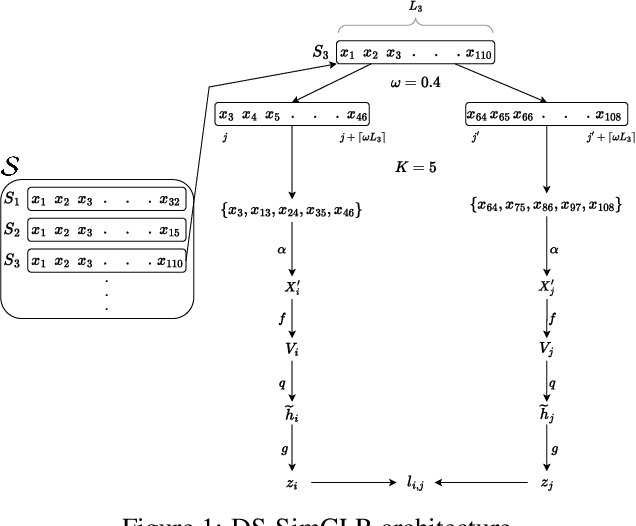
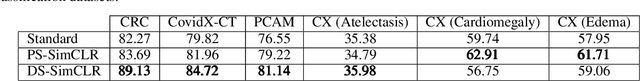
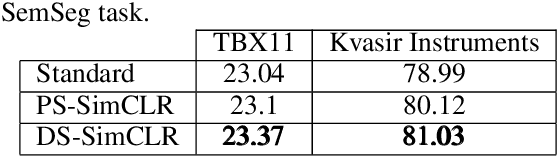
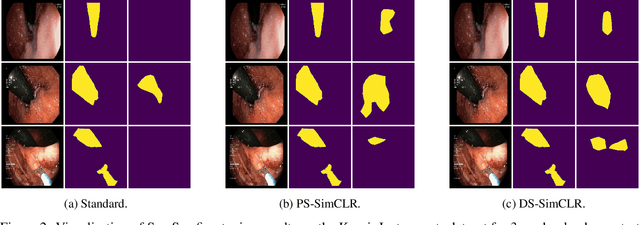
Often, applications of self-supervised learning to 3D medical data opt to use 3D variants of successful 2D network architectures. Although promising approaches, they are significantly more computationally demanding to train, and thus reduce the widespread applicability of these methods away from those with modest computational resources. Thus, in this paper, we aim to improve standard 2D SSL algorithms by modelling the inherent 3D nature of these datasets implicitly. We propose two variants that build upon a strong baseline model and show that both of these variants often outperform the baseline in a variety of downstream tasks. Importantly, in contrast to previous works in both 2D and 3D approaches for 3D medical data, both of our proposals introduce negligible additional overhead over the baseline, improving the democratisation of these approaches for medical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge