Deep learning-based synthetic CT generation from MR images: comparison of generative adversarial and residual neural networks
Paper and Code
Mar 02, 2021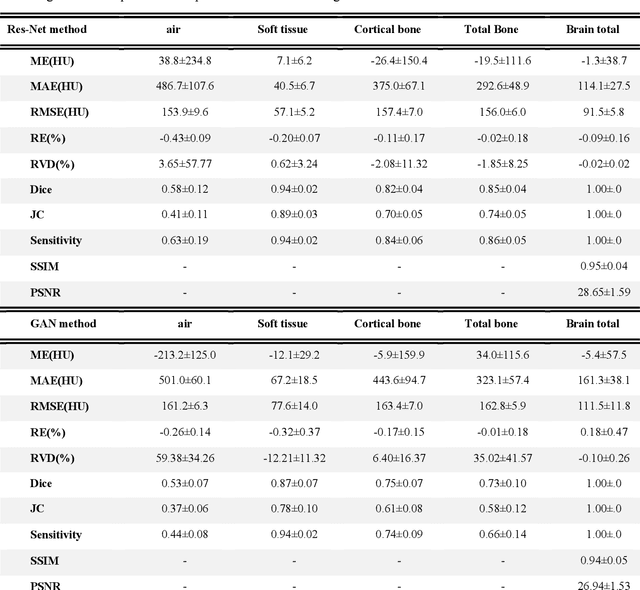
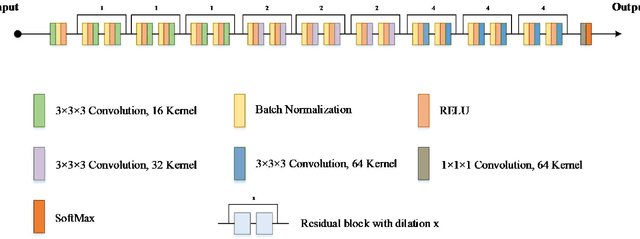
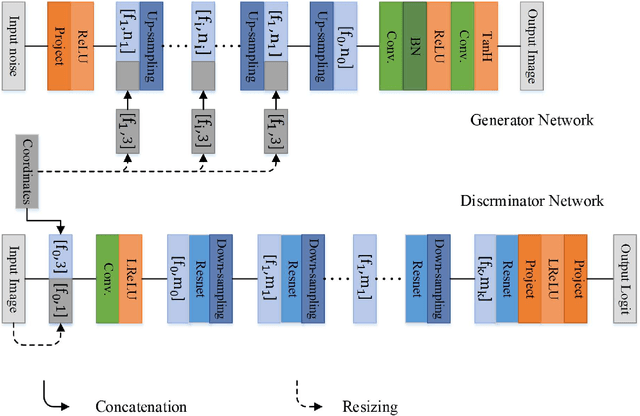
Currently, MRI-only radiotherapy (RT) eliminates some of the concerns about using CT images in RT chains such as the registration of MR images to a separate CT, extra dose delivery, and the additional cost of repeated imaging. However, one remaining challenge is that the signal intensities of MRI are not related to the attenuation coefficient of the biological tissue. This work compares the performance of two state-of-the-art deep learning models; a generative adversarial network (GAN) and a residual network (ResNet) for synthetic CTs (sCT) generation from MR images. The brain MR and CT images of 86 participants were analyzed. GAN and ResNet models were implemented for the generation of synthetic CTs from the 3D T1-weighted MR images using a six-fold cross-validation scheme. The resulting sCTs were compared, considering the CT images as a reference using standard metrics such as the mean absolute error (MAE), peak signal-to-noise ratio (PSNR) and the structural similarity index (SSIM). Overall, the ResNet model exhibited higher accuracy in relation to the delineation of brain tissues. The ResNet model estimated the CT values for the entire head region with an MAE of 114.1 HU compared to MAE=-10.9 HU obtained from the GAN model. Moreover, both models offered comparable SSIM and PSNR values, although the ResNet method exhibited a slightly superior performance over the GAN method. We compared two state-of-the-art deep learning models for the task of MR-based sCT generation. The ResNet model exhibited superior results, thus demonstrating its potential to be used for the challenge of synthetic CT generation in PET/MR AC and MR-only RT planning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge