Deep convolutional neural networks for segmenting 3D in vivo multiphoton images of vasculature in Alzheimer disease mouse models
Paper and Code
Oct 17, 2018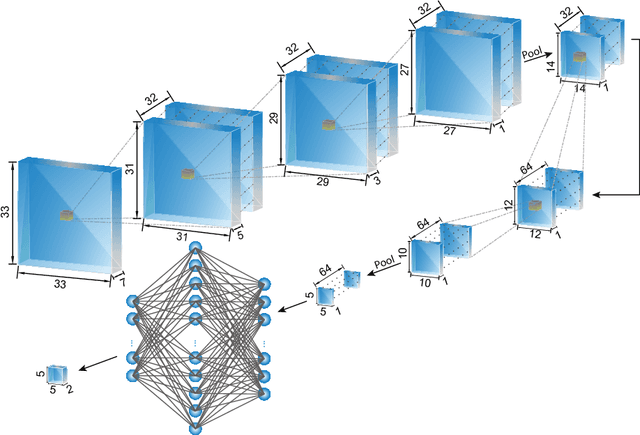
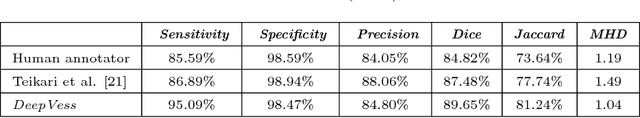


The health and function of tissue rely on its vasculature network to provide reliable blood perfusion. Volumetric imaging approaches, such as multiphoton microscopy, are able to generate detailed 3D images of blood vessels that could contribute to our understanding of the role of vascular structure in normal physiology and in disease mechanisms. The segmentation of vessels, a core image analysis problem, is a bottleneck that has prevented the systematic comparison of 3D vascular architecture across experimental populations. We explored the use of convolutional neural networks to segment 3D vessels within volumetric in vivo images acquired by multiphoton microscopy. We evaluated different network architectures and machine learning techniques in the context of this segmentation problem. We show that our optimized convolutional neural network architecture, which we call DeepVess, yielded a segmentation accuracy that was better than both the current state-of-the-art and a trained human annotator, while also being orders of magnitude faster. To explore the effects of aging and Alzheimer's disease on capillaries, we applied DeepVess to 3D images of cortical blood vessels in young and old mouse models of Alzheimer's disease and wild type littermates. We found little difference in the distribution of capillary diameter or tortuosity between these groups, but did note a decrease in the number of longer capillary segments ($>75\mu m$) in aged animals as compared to young, in both wild type and Alzheimer's disease mouse models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge