Data-Driven Learning of Boolean Networks and Functions by Optimal Causation Entropy Principle
Paper and Code
Jun 01, 2020

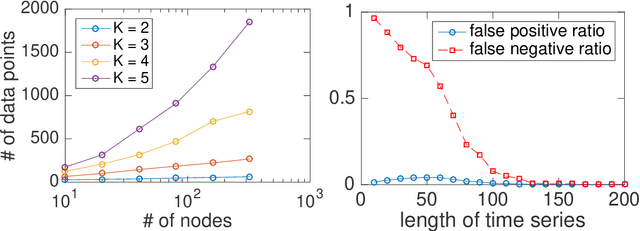
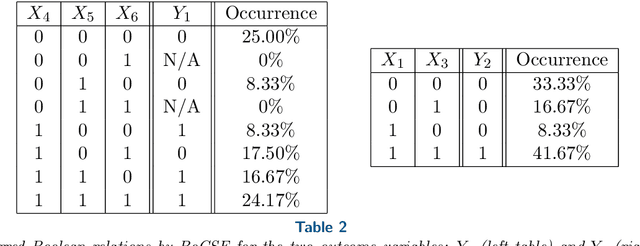
Boolean functions and networks are commonly used in the modeling and analysis of complex biological systems, and this paradigm is highly relevant in other important areas in data science and decision making, such as in the medical field and in the finance industry. Automated learning of a Boolean network and Boolean functions, from data, is a challenging task due in part to the large number of unknowns (including both the structure of the network and the functions) to be estimated, for which a brute force approach would be exponentially complex. In this paper we develop a new information theoretic methodology that we show to be significantly more efficient than previous approaches. Building on the recently developed optimal causation entropy principle (oCSE), that we proved can correctly infer networks distinguishing between direct versus indirect connections, we develop here an efficient algorithm that furthermore infers a Boolean network (including both its structure and function) based on data observed from the evolving states at nodes. We call this new inference method, Boolean optimal causation entropy (BoCSE), which we will show that our method is both computationally efficient and also resilient to noise. Furthermore, it allows for selection of a set of features that best explains the process, a statement that can be described as a networked Boolean function reduced order model. We highlight our method to the feature selection in several real-world examples: (1) diagnosis of urinary diseases, (2) Cardiac SPECT diagnosis, (3) informative positions in the game Tic-Tac-Toe, and (4) risk causality analysis of loans in default status. Our proposed method is effective and efficient in all examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge