Contagion Effect Estimation Using Proximal Embeddings
Paper and Code
Jun 04, 2023
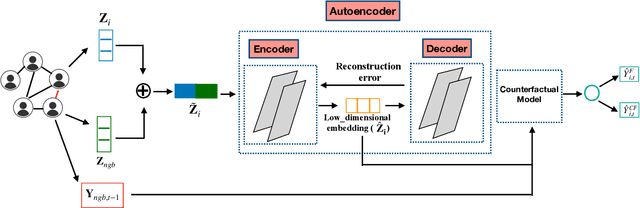
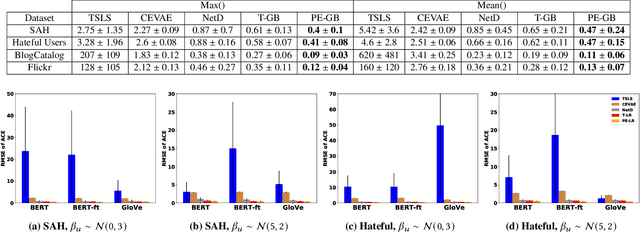

Contagion effect refers to the causal effect of peers' behavior on the outcome of an individual in social networks. While prominent methods for estimating contagion effects in observational studies often assume that there are no unmeasured confounders, contagion can be confounded due to latent homophily: nodes in a homophilous network tend to have ties to peers with similar attributes and can behave similarly without influencing one another. One way to account for latent homophily is by considering proxies for the unobserved confounders. However, in the presence of high-dimensional proxies, proxy-based methods can lead to substantially biased estimation of contagion effects, as we demonstrate in this paper. To tackle this issue, we introduce the novel Proximal Embeddings (ProEmb), a framework which integrates Variational Autoencoders (VAEs) and adversarial networks to generate balanced low-dimensional representations of high-dimensional proxies for different treatment groups and identifies contagion effects in the presence of unobserved network confounders. We empirically show that our method significantly increases the accuracy of contagion effect estimation in observational network data compared to state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge