Community Detection and Classification in Hierarchical Stochastic Blockmodels
Paper and Code
Aug 26, 2016
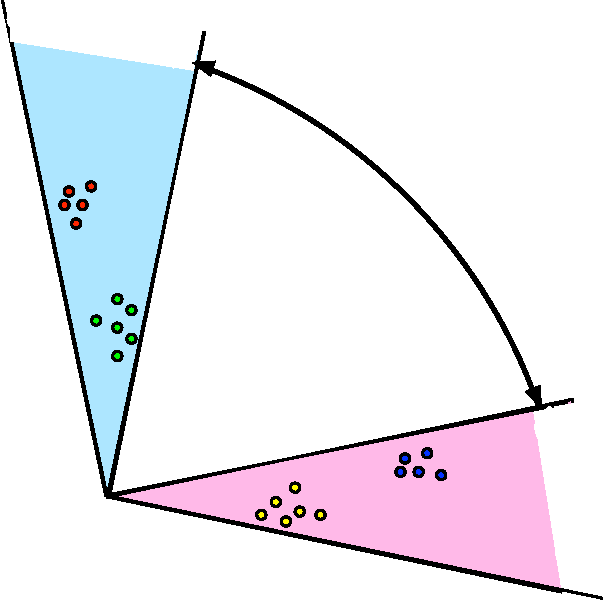
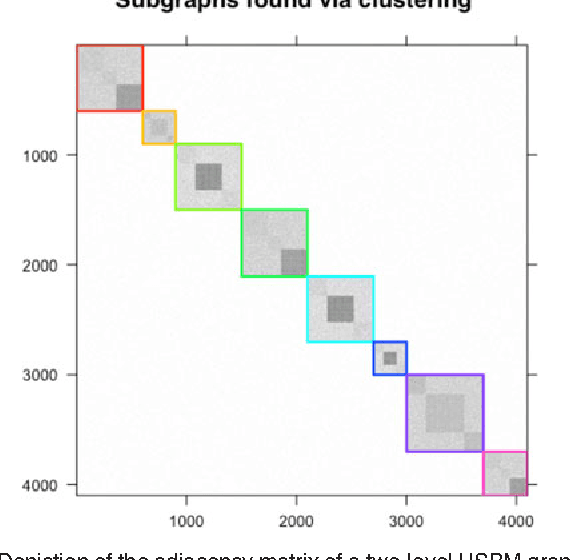
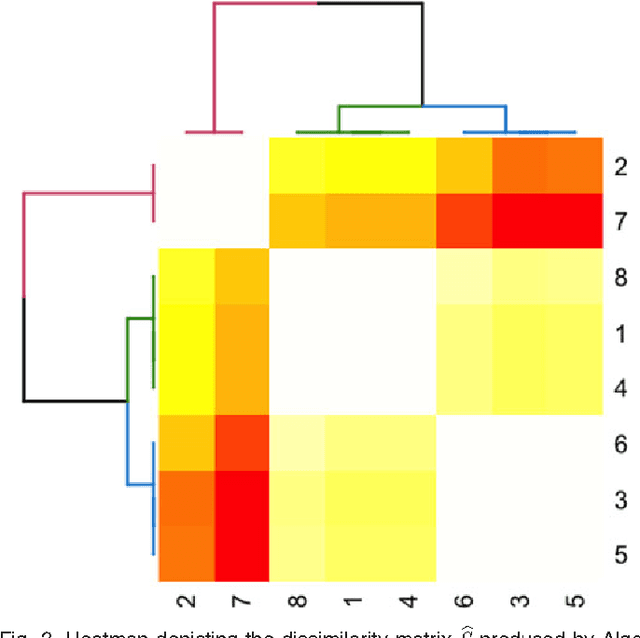
We propose a robust, scalable, integrated methodology for community detection and community comparison in graphs. In our procedure, we first embed a graph into an appropriate Euclidean space to obtain a low-dimensional representation, and then cluster the vertices into communities. We next employ nonparametric graph inference techniques to identify structural similarity among these communities. These two steps are then applied recursively on the communities, allowing us to detect more fine-grained structure. We describe a hierarchical stochastic blockmodel---namely, a stochastic blockmodel with a natural hierarchical structure---and establish conditions under which our algorithm yields consistent estimates of model parameters and motifs, which we define to be stochastically similar groups of subgraphs. Finally, we demonstrate the effectiveness of our algorithm in both simulated and real data. Specifically, we address the problem of locating similar subcommunities in a partially reconstructed Drosophila connectome and in the social network Friendster.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge