Combining haplotypers
Paper and Code
Oct 26, 2007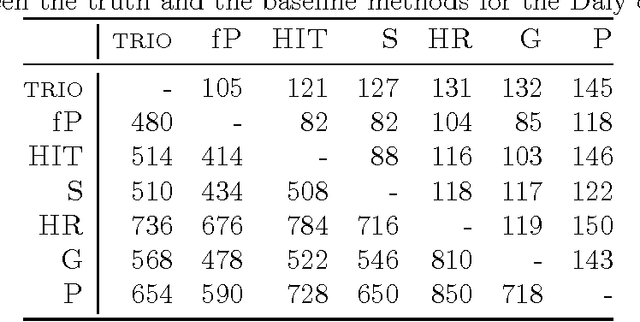
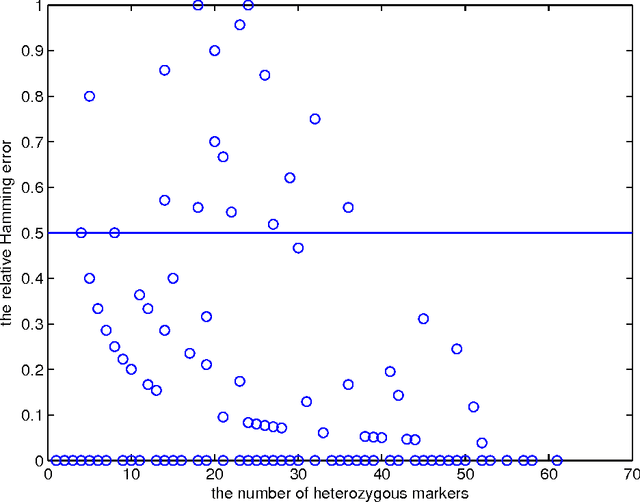
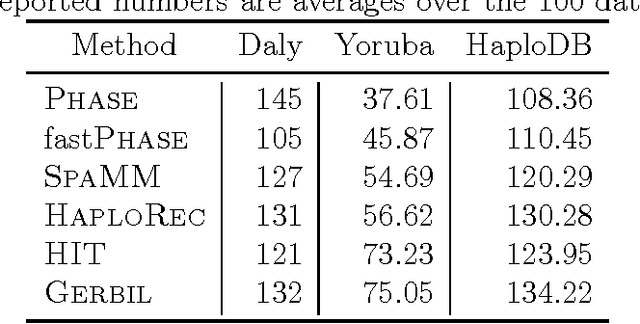
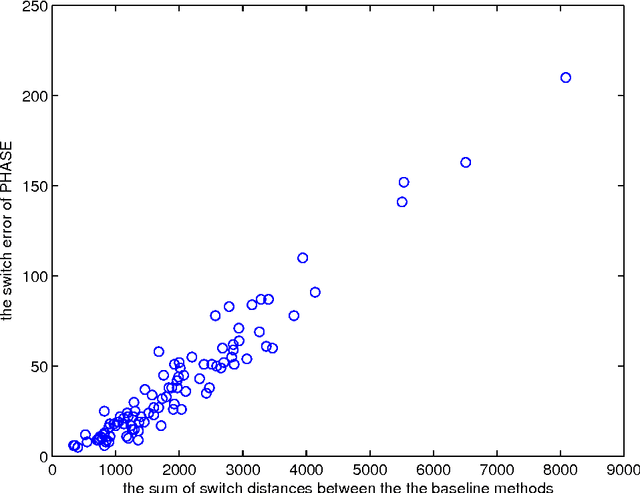
Statistically resolving the underlying haplotype pair for a genotype measurement is an important intermediate step in gene mapping studies, and has received much attention recently. Consequently, a variety of methods for this problem have been developed. Different methods employ different statistical models, and thus implicitly encode different assumptions about the nature of the underlying haplotype structure. Depending on the population sample in question, their relative performance can vary greatly, and it is unclear which method to choose for a particular sample. Instead of choosing a single method, we explore combining predictions returned by different methods in a principled way, and thereby circumvent the problem of method selection. We propose several techniques for combining haplotype reconstructions and analyze their computational properties. In an experimental study on real-world haplotype data we show that such techniques can provide more accurate and robust reconstructions, and are useful for outlier detection. Typically, the combined prediction is at least as accurate as or even more accurate than the best individual method, effectively circumventing the method selection problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge