Clustering Propagation for Universal Medical Image Segmentation
Paper and Code
Mar 25, 2024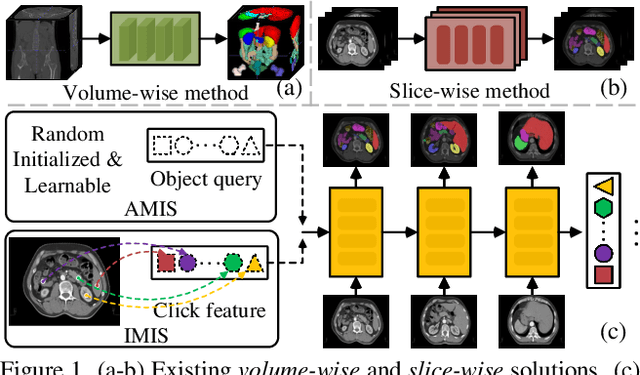
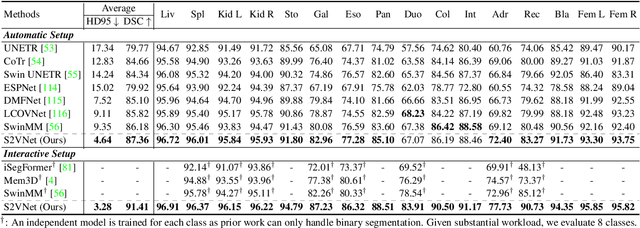
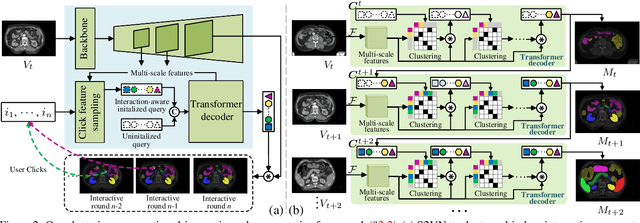
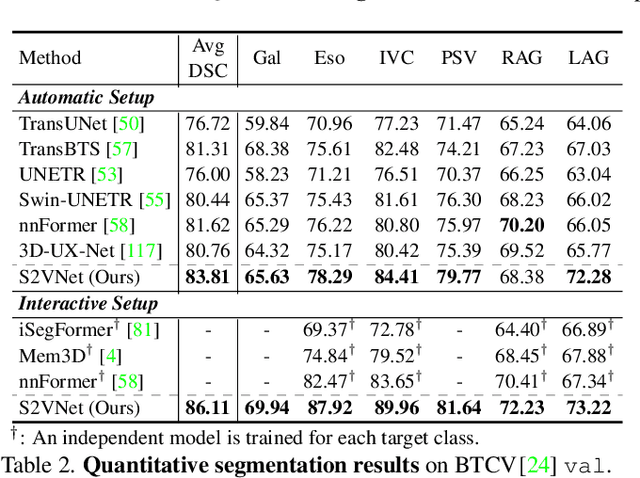
Prominent solutions for medical image segmentation are typically tailored for automatic or interactive setups, posing challenges in facilitating progress achieved in one task to another.$_{\!}$ This$_{\!}$ also$_{\!}$ necessitates$_{\!}$ separate$_{\!}$ models for each task, duplicating both training time and parameters.$_{\!}$ To$_{\!}$ address$_{\!}$ above$_{\!}$ issues,$_{\!}$ we$_{\!}$ introduce$_{\!}$ S2VNet,$_{\!}$ a$_{\!}$ universal$_{\!}$ framework$_{\!}$ that$_{\!}$ leverages$_{\!}$ Slice-to-Volume$_{\!}$ propagation$_{\!}$ to$_{\!}$ unify automatic/interactive segmentation within a single model and one training session. Inspired by clustering-based segmentation techniques, S2VNet makes full use of the slice-wise structure of volumetric data by initializing cluster centers from the cluster$_{\!}$ results$_{\!}$ of$_{\!}$ previous$_{\!}$ slice.$_{\!}$ This enables knowledge acquired from prior slices to assist in the segmentation of the current slice, further efficiently bridging the communication between remote slices using mere 2D networks. Moreover, such a framework readily accommodates interactive segmentation with no architectural change, simply by initializing centroids from user inputs. S2VNet distinguishes itself by swift inference speeds and reduced memory consumption compared to prevailing 3D solutions. It can also handle multi-class interactions with each of them serving to initialize different centroids. Experiments on three benchmarks demonstrate S2VNet surpasses task-specified solutions on both automatic/interactive setups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge