Clustering and classification of low-dimensional data in explicit feature map domain: intraoperative pixel-wise diagnosis of adenocarcinoma of a colon in a liver
Paper and Code
Mar 07, 2022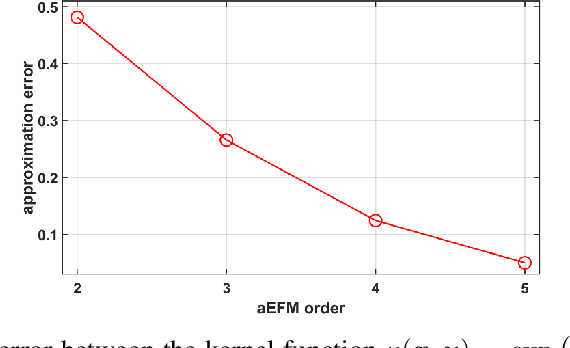
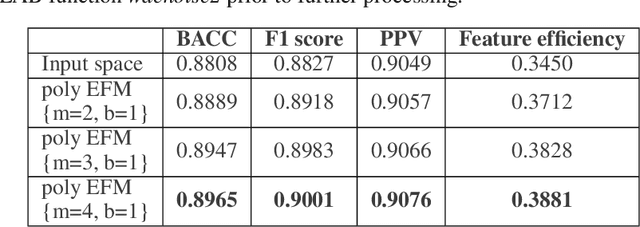
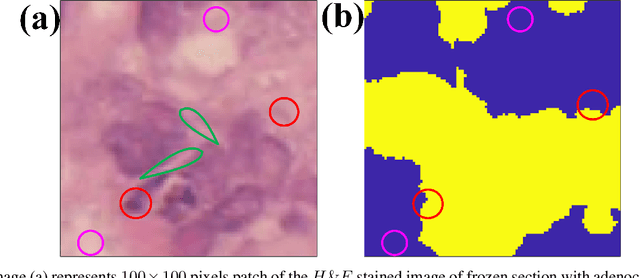
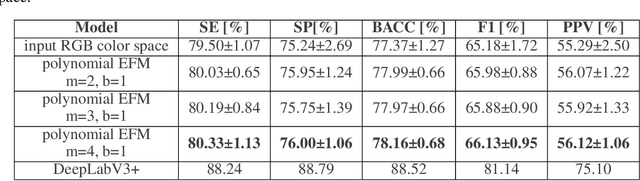
Application of artificial intelligence in medicine brings in highly accurate predictions achieved by complex models, the reasoning of which is hard to interpret. Their generalization ability can be reduced because of the lack of pixel wise annotated images that occurs in frozen section tissue analysis. To partially overcome this gap, this paper explores the approximate explicit feature map (aEFM) transform of low-dimensional data into a low-dimensional subspace in Hilbert space. There, with a modest increase in computational complexity, linear algorithms yield improved performance and keep interpretability. They remain amenable to incremental learning that is not a trivial issue for some nonlinear algorithms. We demonstrate proposed methodology on a very large-scale problem related to intraoperative pixel-wise semantic segmentation and clustering of adenocarcinoma of a colon in a liver. Compared to the results in the input space, logistic classifier achieved statistically significant performance improvements in micro balanced accuracy and F1 score in the amounts of 12.04% and 12.58%, respectively. Support vector machine classifier yielded the increase of 8.04% and 9.41%. For clustering, increases of 0.79% and 0.85% are obtained with ultra large-scale spectral clustering algorithm. Results are supported by a discussion of interpretability using Shapely additive explanation values for predictions of linear classifier in input space and aEFM induced space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge