Building Models for Biopathway Dynamics Using Intrinsic Dimensionality Analysis
Paper and Code
Nov 03, 2018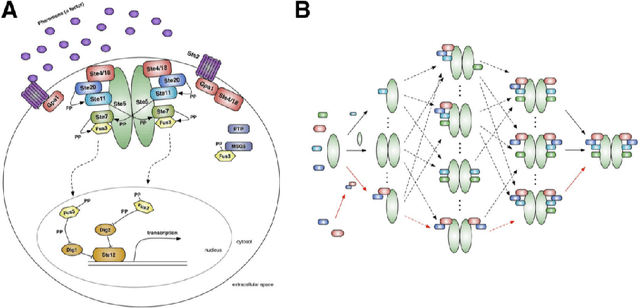
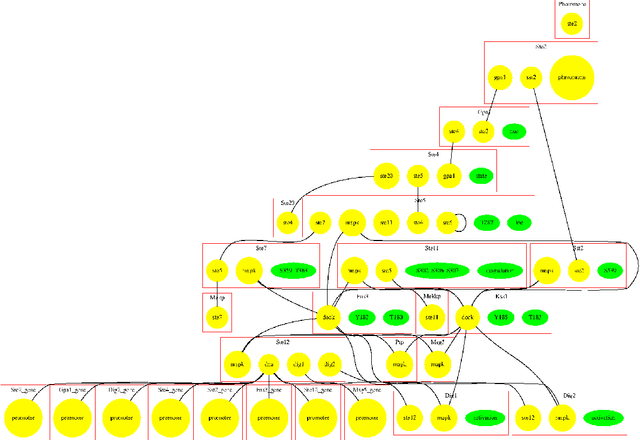
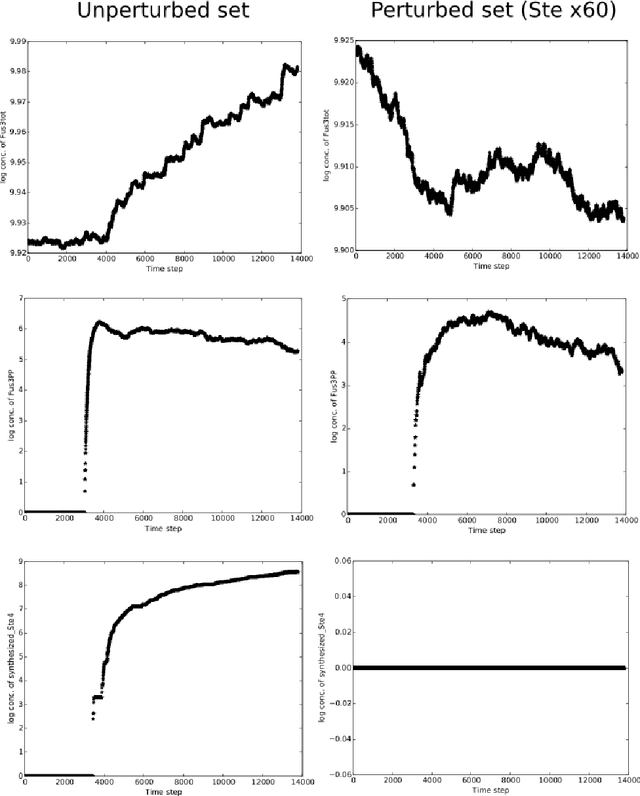
An important task for many if not all the scientific domains is efficient knowledge integration, testing and codification. It is often solved with model construction in a controllable computational environment. In spite of that, the throughput of in-silico simulation-based observations become similarly intractable for thorough analysis. This is especially the case in molecular biology, which served as a subject for this study. In this project, we aimed to test some approaches developed to deal with the curse of dimensionality. Among these we found dimension reduction techniques especially appealing. They can be used to identify irrelevant variability and help to understand critical processes underlying high-dimensional datasets. Additionally, we subjected our data sets to nonlinear time series analysis, as those are well established methods for results comparison. To investigate the usefulness of dimension reduction methods, we decided to base our study on a concrete sample set. The example was taken from the domain of systems biology concerning dynamic evolution of sub-cellular signaling. Particularly, the dataset relates to the yeast pheromone pathway and is studied in-silico with a stochastic model. The model reconstructs signal propagation stimulated by a mating pheromone. In the paper, we elaborate on the reason of multidimensional analysis problem in the context of molecular signaling, and next, we introduce the model of choice, simulation details and obtained time series dynamics. A description of used methods followed by a discussion of results and their biological interpretation finalize the paper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge