BROW: Better featuRes fOr Whole slide image based on self-distillation
Paper and Code
Sep 15, 2023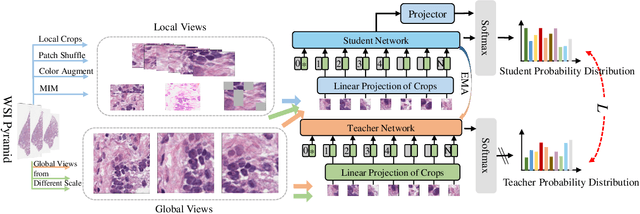
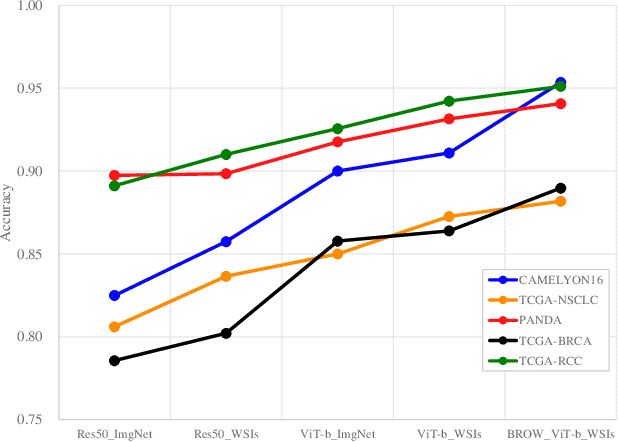
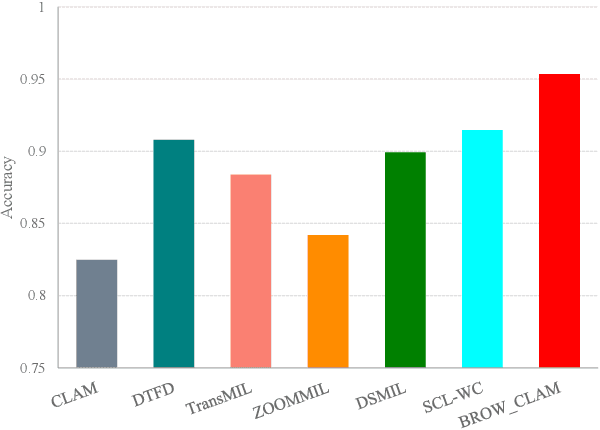
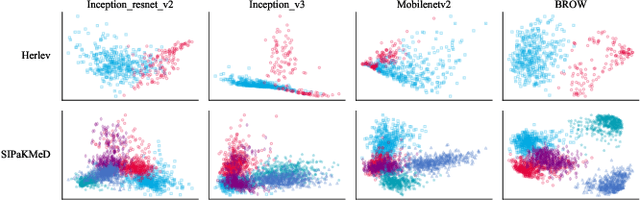
Whole slide image (WSI) processing is becoming part of the key components of standard clinical diagnosis for various diseases. However, the direct application of conventional image processing algorithms to WSI faces certain obstacles because of WSIs' distinct property: the super-high resolution. The performance of most WSI-related tasks relies on the efficacy of the backbone which extracts WSI patch feature representations. Hence, we proposed BROW, a foundation model for extracting better feature representations for WSIs, which can be conveniently adapted to downstream tasks without or with slight fine-tuning. The model takes transformer architecture, pretrained using self-distillation framework. To improve model's robustness, techniques such as patch shuffling have been employed. Additionally, the model leverages the unique properties of WSIs, utilizing WSI's multi-scale pyramid to incorporate an additional global view, thereby further enhancing its performance. We used both private and public data to make up a large pretraining dataset, containing more than 11000 slides, over 180M extracted patches, encompassing WSIs related to various organs and tissues. To assess the effectiveness of \ourmodel, we run a wide range of downstream tasks, including slide-level subtyping, patch-level classification and nuclei instance segmentation. The results confirmed the efficacy, robustness and good generalization ability of the proposed model. This substantiates its potential as foundation model for WSI feature extraction and highlights promising prospects for its application in WSI processing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge