Behavioral-clinical phenotyping with type 2 diabetes self-monitoring data
Paper and Code
Feb 23, 2018
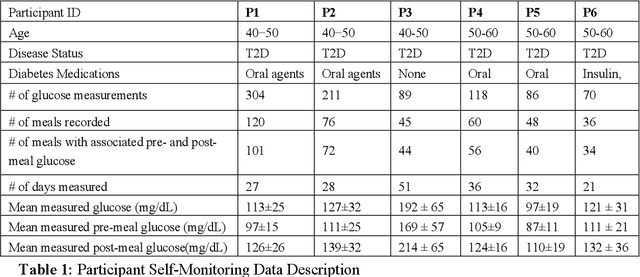
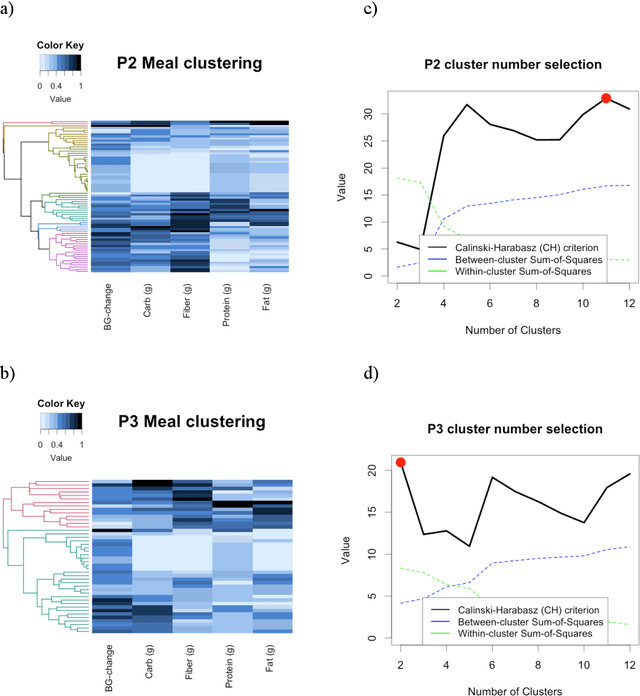

Objective: To evaluate unsupervised clustering methods for identifying individual-level behavioral-clinical phenotypes that relate personal biomarkers and behavioral traits in type 2 diabetes (T2DM) self-monitoring data. Materials and Methods: We used hierarchical clustering (HC) to identify groups of meals with similar nutrition and glycemic impact for 6 individuals with T2DM who collected self-monitoring data. We evaluated clusters on: 1) correspondence to gold standards generated by certified diabetes educators (CDEs) for 3 participants; 2) face validity, rated by CDEs, and 3) impact on CDEs' ability to identify patterns for another 3 participants. Results: Gold standard (GS) included 9 patterns across 3 participants. Of these, all 9 were re-discovered using HC: 4 GS patterns were consistent with patterns identified by HC (over 50% of meals in a cluster followed the pattern); another 5 were included as sub-groups in broader clusers. 50% (9/18) of clusters were rated over 3 on 5-point Likert scale for validity, significance, and being actionable. After reviewing clusters, CDEs identified patterns that were more consistent with data (70% reduction in contradictions between patterns and participants' records). Discussion: Hierarchical clustering of blood glucose and macronutrient consumption appears suitable for discovering behavioral-clinical phenotypes in T2DM. Most clusters corresponded to gold standard and were rated positively by CDEs for face validity. Cluster visualizations helped CDEs identify more robust patterns in nutrition and glycemic impact, creating new possibilities for visual analytic solutions. Conclusion: Machine learning methods can use diabetes self-monitoring data to create personalized behavioral-clinical phenotypes, which may prove useful for delivering personalized medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge