Augmented Transformer Achieves 97% and 85% for Top5 Prediction of Direct and Classical Retro-Synthesis
Paper and Code
Mar 05, 2020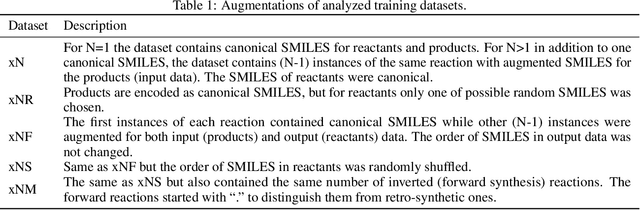
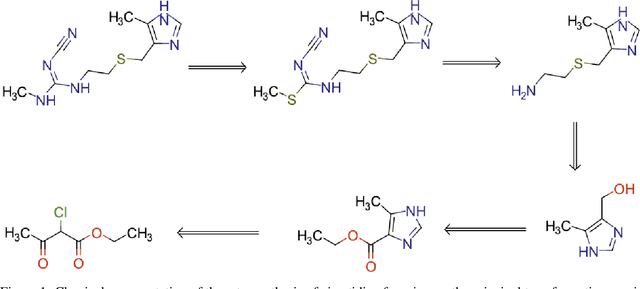
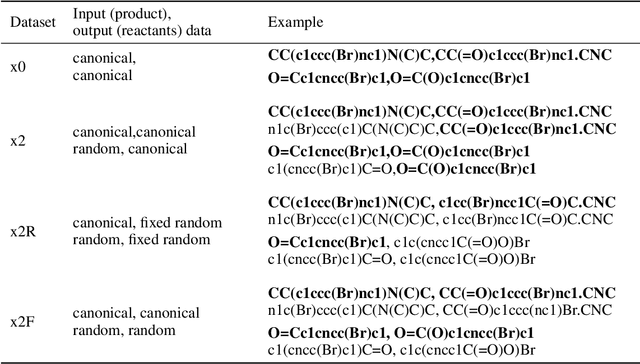
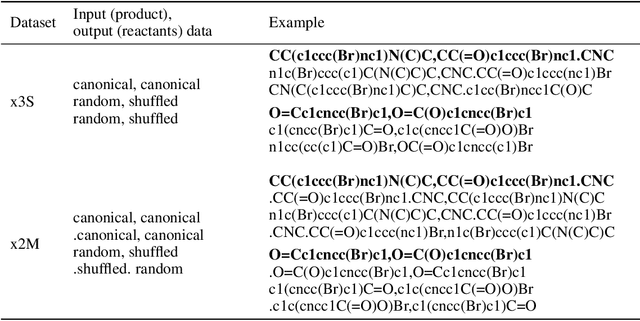
We investigated the effect of different augmentation scenarios on predicting (retro)synthesis of chemical compounds using SMILES representation. We showed that augmentation of not only input sequences but also, importantly, of the target data eliminated the effect of data memorization by neural networks and improved their generalization performance for prediction of new sequences. The Top-5 accuracy was 85.4% for the prediction of the largest fragment (thus identifying principal transformation for classical retro-synthesis) for USPTO-50k test dataset and was achieved by a combination of SMILES augmentation and beam search. The same approach also outperformed best published results for prediction of direct reactions from the USPTO-MIT test set. Our model achieved 90.4% Top-1 and 96.5% Top-5 accuracy for its most challenging mixed set and 97% Top-5 accuracy for the USPTO-MIT separated set. The appearance frequency of the most abundantly generated SMILES was well correlated with the prediction outcome and can be used as a measure of the quality of reaction prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge