An SIR Graph Growth Model for the Epidemics of Communicable Diseases
Paper and Code
Feb 05, 2014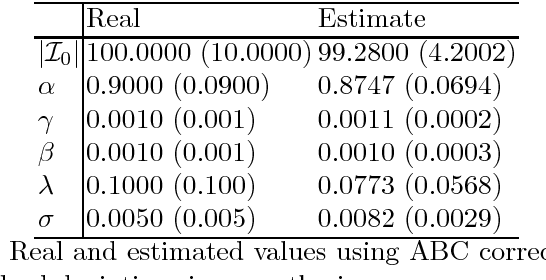
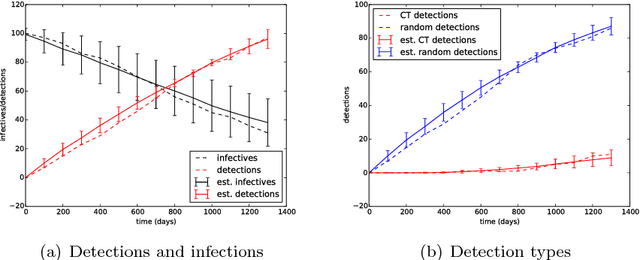
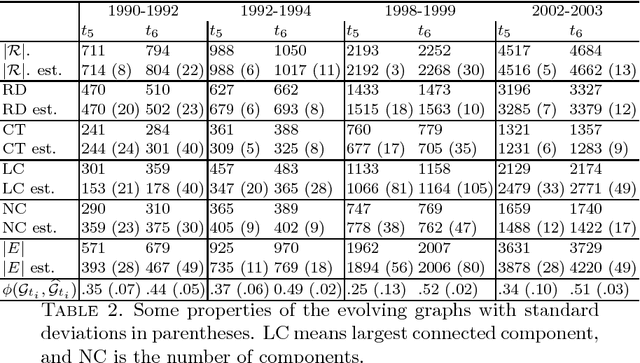
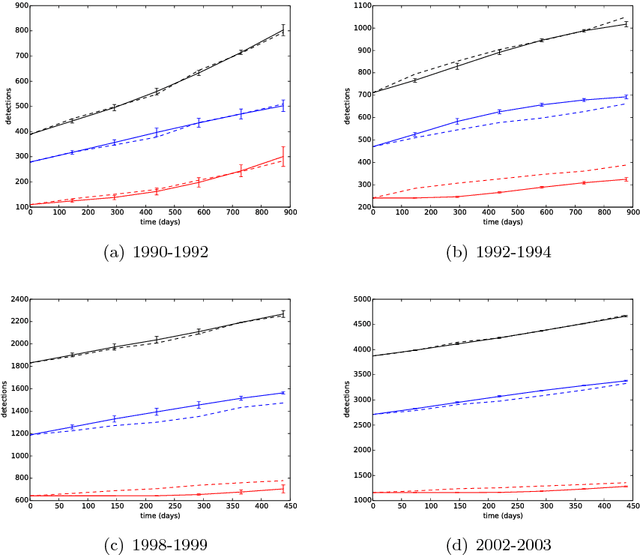
It is the main purpose of this paper to introduce a graph-valued stochastic process in order to model the spread of a communicable infectious disease. The major novelty of the SIR model we promote lies in the fact that the social network on which the epidemics is taking place is not specified in advance but evolves through time, accounting for the temporal evolution of the interactions involving infective individuals. Without assuming the existence of a fixed underlying network model, the stochastic process introduced describes, in a flexible and realistic manner, epidemic spread in non-uniformly mixing and possibly heterogeneous populations. It is shown how to fit such a (parametrised) model by means of Approximate Bayesian Computation methods based on graph-valued statistics. The concepts and statistical methods described in this paper are finally applied to a real epidemic dataset, related to the spread of HIV in Cuba in presence of a contact tracing system, which permits one to reconstruct partly the evolution of the graph of sexual partners diagnosed HIV positive between 1986 and 2006.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge