An Efficient Training Algorithm for Kernel Survival Support Vector Machines
Paper and Code
Nov 21, 2016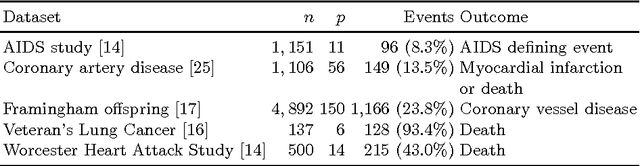
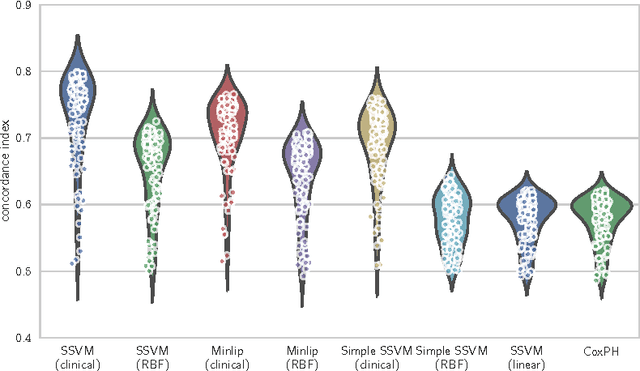
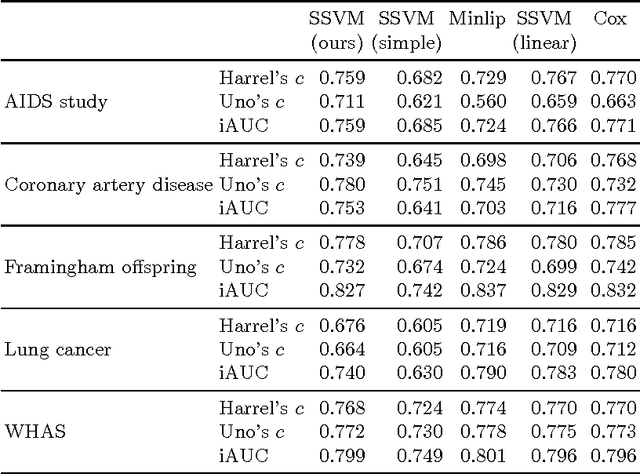
Survival analysis is a fundamental tool in medical research to identify predictors of adverse events and develop systems for clinical decision support. In order to leverage large amounts of patient data, efficient optimisation routines are paramount. We propose an efficient training algorithm for the kernel survival support vector machine (SSVM). We directly optimise the primal objective function and employ truncated Newton optimisation and order statistic trees to significantly lower computational costs compared to previous training algorithms, which require $O(n^4)$ space and $O(p n^6)$ time for datasets with $n$ samples and $p$ features. Our results demonstrate that our proposed optimisation scheme allows analysing data of a much larger scale with no loss in prediction performance. Experiments on synthetic and 5 real-world datasets show that our technique outperforms existing kernel SSVM formulations if the amount of right censoring is high ($\geq85\%$), and performs comparably otherwise.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge