A Unified View of Stochastic Hamiltonian Sampling
Paper and Code
Jun 30, 2021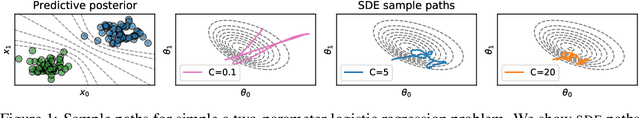
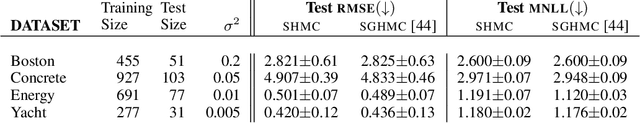
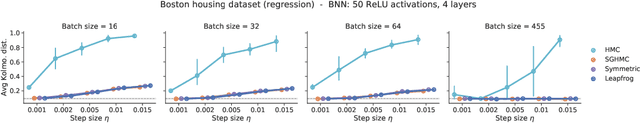
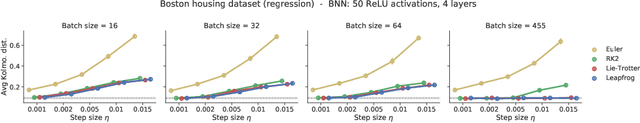
In this work, we revisit the theoretical properties of Hamiltonian stochastic differential equations (SDEs) for Bayesian posterior sampling, and we study the two types of errors that arise from numerical SDE simulation: the discretization error and the error due to noisy gradient estimates in the context of data subsampling. We consider overlooked results describing the ergodic convergence rates of numerical integration schemes, and we produce a novel analysis for the effect of mini-batches through the lens of differential operator splitting. In our analysis, the stochastic component of the proposed Hamiltonian SDE is decoupled from the gradient noise, for which we make no normality assumptions. This allows us to derive interesting connections among different sampling schemes, including the original Hamiltonian Monte Carlo (HMC) algorithm, and explain their performance. We show that for a careful selection of numerical integrators, both errors vanish at a rate $\mathcal{O}(\eta^2)$, where $\eta$ is the integrator step size. Our theoretical results are supported by an empirical study on a variety of regression and classification tasks for Bayesian neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge