A Note on Optimal Sampling Strategy for Structural Variant Detection Using Optical Mapping
Paper and Code
Oct 04, 2019
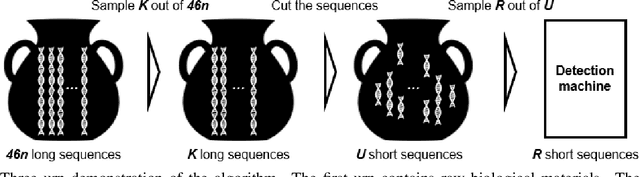
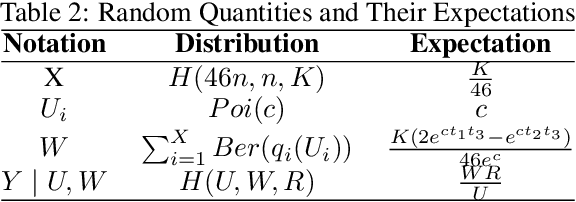

Structural variants compose the majority of human genetic variation, but are difficult to assess using current genomic sequencing technologies. Optical mapping technologies, which measure the size of chromosomal fragments between labeled markers, offer an alternative approach. As these technologies mature towards becoming clinical tools, there is a need to develop an approach for determining the optimal strategy for sampling biological material in order to detect a variant at some threshold. Here we develop an optimization approach using a simple, yet realistic, model of the genomic mapping process using a hyper-geometric distribution and {probabilistic} concentration inequalities. Our approach is both computationally and analytically tractable and includes a novel approach to getting tail bounds of hyper-geometric distribution. We show that if a genomic mapping technology can sample most of the chromosomal fragments within a sample, comparatively little biological material is needed to detect a variant at high confidence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge