A new nature inspired modularity function adapted for unsupervised learning involving spatially embedded networks: A comparative analysis
Paper and Code
Jul 18, 2020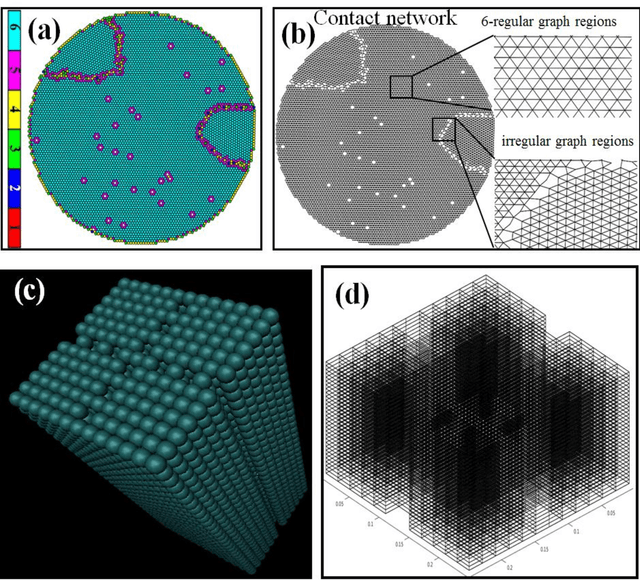
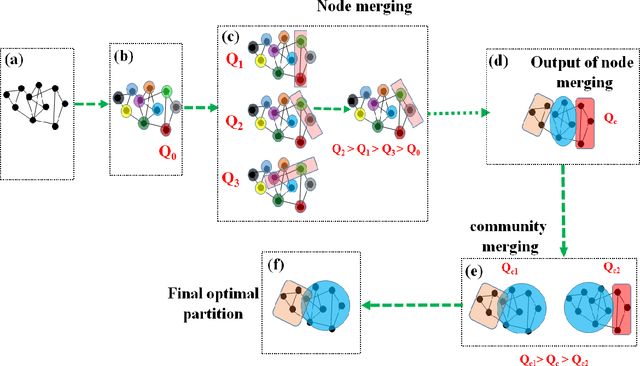
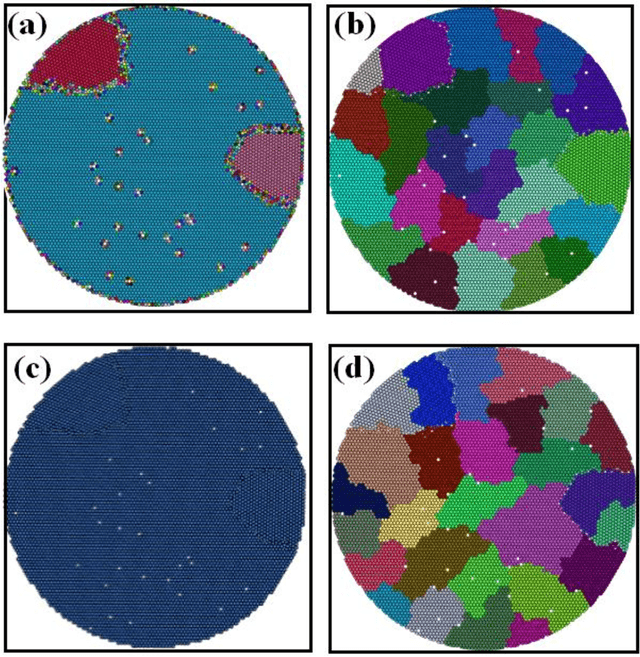
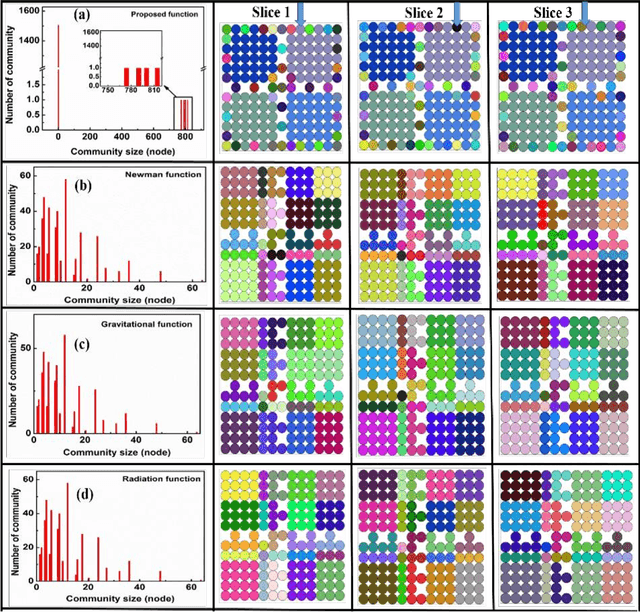
Unsupervised machine learning methods can be of great help in many traditional engineering disciplines, where huge amount of labeled data is not readily available or is extremely difficult or costly to generate. Two specific examples include the structure of granular materials and atomic structure of metallic glasses. While the former is critically important for several hundreds of billion dollars global industries, the latter is still a big puzzle in fundamental science. One thing is common in both the examples is that the particles are the elements of the ensembles that are embedded in Euclidean space and one can create a spatially embedded network to represent their key features. Some recent studies show that clustering, which generically refers to unsupervised learning, holds great promise in partitioning these networks. In many complex networks, the spatial information of nodes play very important role in determining the network properties. So understanding the structure of such networks is very crucial. We have compared the performance of our newly developed modularity function with some of the well-known modularity functions. We performed this comparison by finding the best partition in 2D and 3D granular assemblies. We show that for the class of networks considered in this article, our method produce much better results than the competing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge