A Hyperspectral Imaging Dataset and Methodology for Intraoperative Pixel-Wise Classification of Metastatic Colon Cancer in the Liver
Paper and Code
Nov 11, 2024
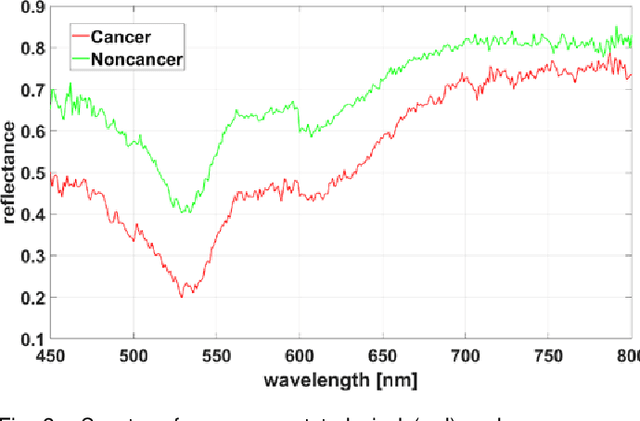
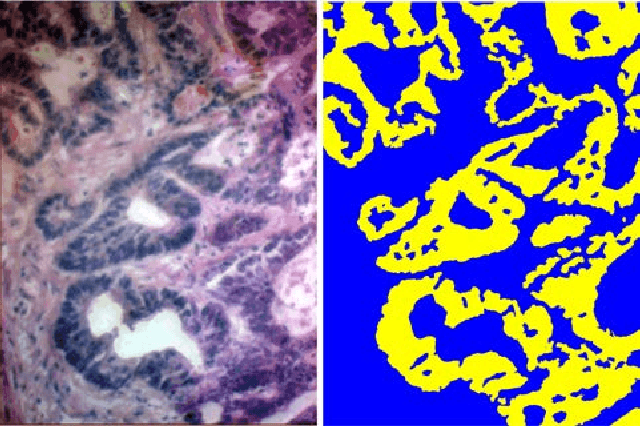
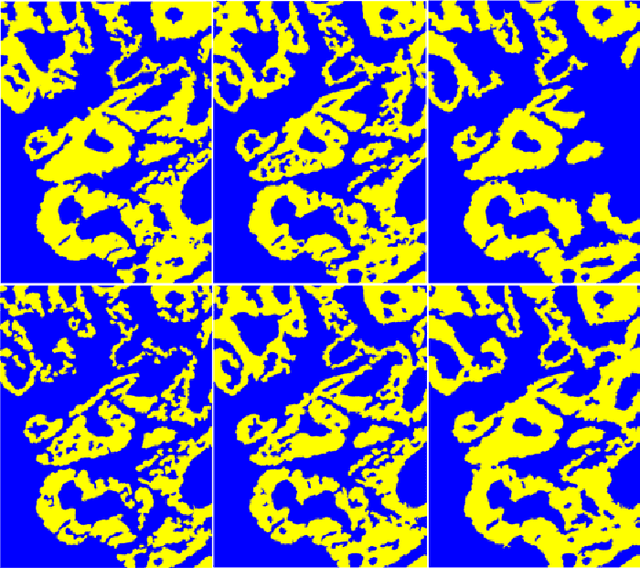
Hyperspectral imaging (HSI) holds significant potential for transforming the field of computational pathology. However, there is currently a shortage of pixel-wise annotated HSI data necessary for training deep learning (DL) models. Additionally, the number of HSI-based research studies remains limited, and in many cases, the advantages of HSI over traditional RGB imaging have not been conclusively demonstrated, particularly for specimens collected intraoperatively. To address these challenges we present a database consisted of 27 HSIs of hematoxylin-eosin stained frozen sections, collected from 14 patients with colon adenocarcinoma metastasized to the liver. It is aimed to validate pixel-wise classification for intraoperative tumor resection. The HSIs were acquired in the spectral range of 450 to 800 nm, with a resolution of 1 nm, resulting in images of 1384x1035 pixels. Pixel-wise annotations were performed by three pathologists. To overcome challenges such as experimental variability and the lack of annotated data, we combined label-propagation-based semi-supervised learning (SSL) with spectral-spatial features extracted by: the multiscale principle of relevant information (MPRI) method and tensor singular spectrum analysis method. Using only 1% of labeled pixels per class the SSL-MPRI method achieved a micro balanced accuracy (BACC) of 0.9313 and a micro F1-score of 0.9235 on the HSI dataset. The performance on corresponding RGB images was lower, with a micro BACC of 0.8809 and a micro F1-score of 0.8688. These improvements are statistically significant. The SSL-MPRI approach outperformed six DL architectures trained with 63% of labeled pixels. Data and code are available at: https://github.com/ikopriva/ColonCancerHSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge