A Hierarchical Approach to Scaling Batch Active Search Over Structured Data
Paper and Code
Jul 20, 2020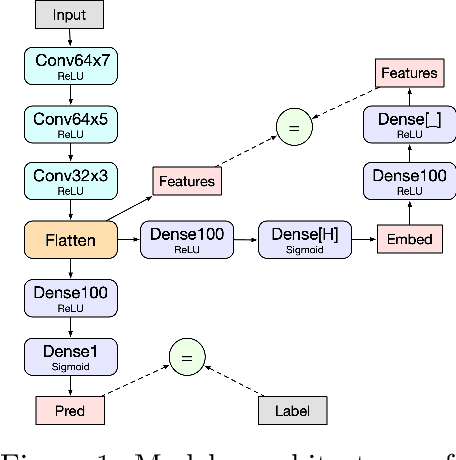
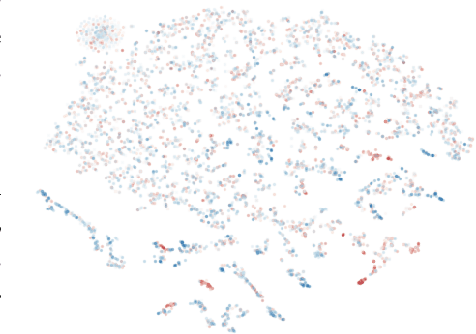
Active search is the process of identifying high-value data points in a large and often high-dimensional parameter space that can be expensive to evaluate. Traditional active search techniques like Bayesian optimization trade off exploration and exploitation over consecutive evaluations, and have historically focused on single or small (<5) numbers of examples evaluated per round. As modern data sets grow, so does the need to scale active search to large data sets and batch sizes. In this paper, we present a general hierarchical framework based on bandit algorithms to scale active search to large batch sizes by maximizing information derived from the unique structure of each dataset. Our hierarchical framework, Hierarchical Batch Bandit Search (HBBS), strategically distributes batch selection across a learned embedding space by facilitating wide exploration of different structural elements within a dataset. We focus our application of HBBS on modern biology, where large batch experimentation is often fundamental to the research process, and demonstrate batch design of biological sequences (protein and DNA). We also present a new Gym environment to easily simulate diverse biological sequences and to enable more comprehensive evaluation of active search methods across heterogeneous data sets. The HBBS framework improves upon standard performance, wall-clock, and scalability benchmarks for batch search by using a broad exploration strategy across coarse partitions and fine-grained exploitation within each partition of structured data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge