A general framework for the IT-based clustering methods
Paper and Code
Jun 19, 2015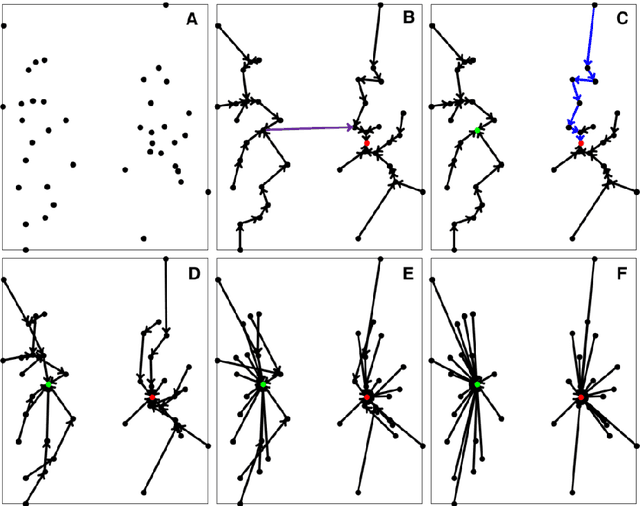
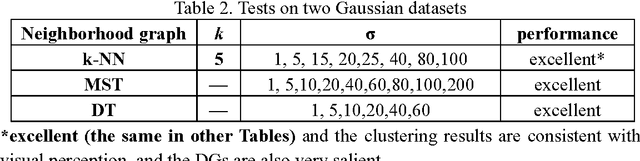
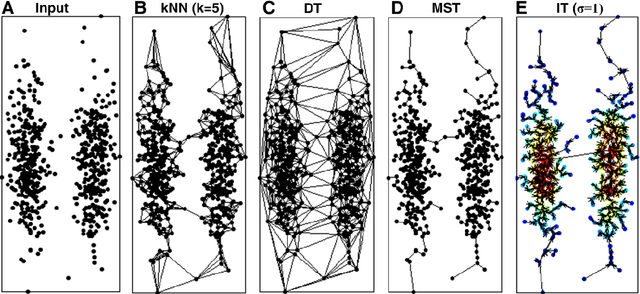
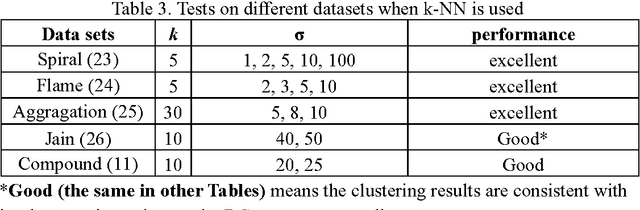
Previously, we proposed a physically inspired rule to organize the data points in a sparse yet effective structure, called the in-tree (IT) graph, which is able to capture a wide class of underlying cluster structures in the datasets, especially for the density-based datasets. Although there are some redundant edges or lines between clusters requiring to be removed by computer, this IT graph has a big advantage compared with the k-nearest-neighborhood (k-NN) or the minimal spanning tree (MST) graph, in that the redundant edges in the IT graph are much more distinguishable and thus can be easily determined by several methods previously proposed by us. In this paper, we propose a general framework to re-construct the IT graph, based on an initial neighborhood graph, such as the k-NN or MST, etc, and the corresponding graph distances. For this general framework, our previous way of constructing the IT graph turns out to be a special case of it. This general framework 1) can make the IT graph capture a wider class of underlying cluster structures in the datasets, especially for the manifolds, and 2) should be more effective to cluster the sparse or graph-based datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge