A Distance Covariance-based Kernel for Nonlinear Causal Clustering in Heterogeneous Populations
Paper and Code
Jun 07, 2021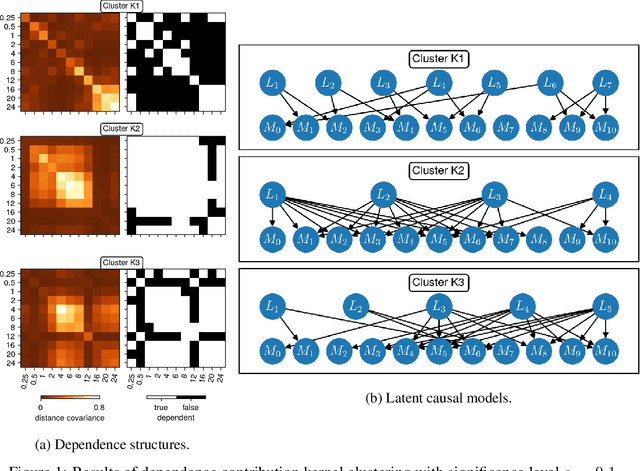
We consider the problem of causal structure learning in the setting of heterogeneous populations, i.e., populations in which a single causal structure does not adequately represent all population members, as is common in biological and social sciences. To this end, we introduce a distance covariance-based kernel designed specifically to measure the similarity between the underlying nonlinear causal structures of different samples. This kernel enables us to perform clustering to identify the homogeneous subpopulations. Indeed, we prove the corresponding feature map is a statistically consistent estimator of nonlinear independence structure, rendering the kernel itself a statistical test for the hypothesis that sets of samples come from different generating causal structures. We can then use existing methods to learn a causal structure for each of these subpopulations. We demonstrate using our kernel for causal clustering with an application in genetics, allowing us to reason about the latent transcription factor networks regulating measured gene expression levels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge