3-Dimensional Deep Learning with Spatial Erasing for Unsupervised Anomaly Segmentation in Brain MRI
Paper and Code
Sep 14, 2021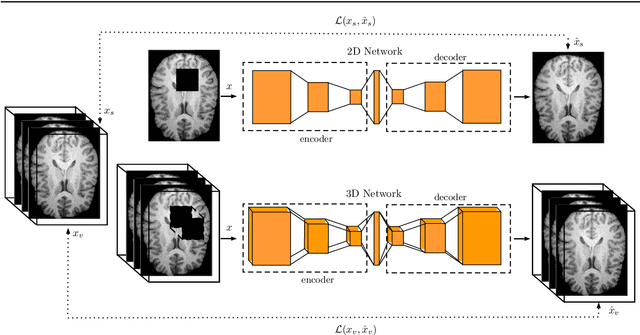
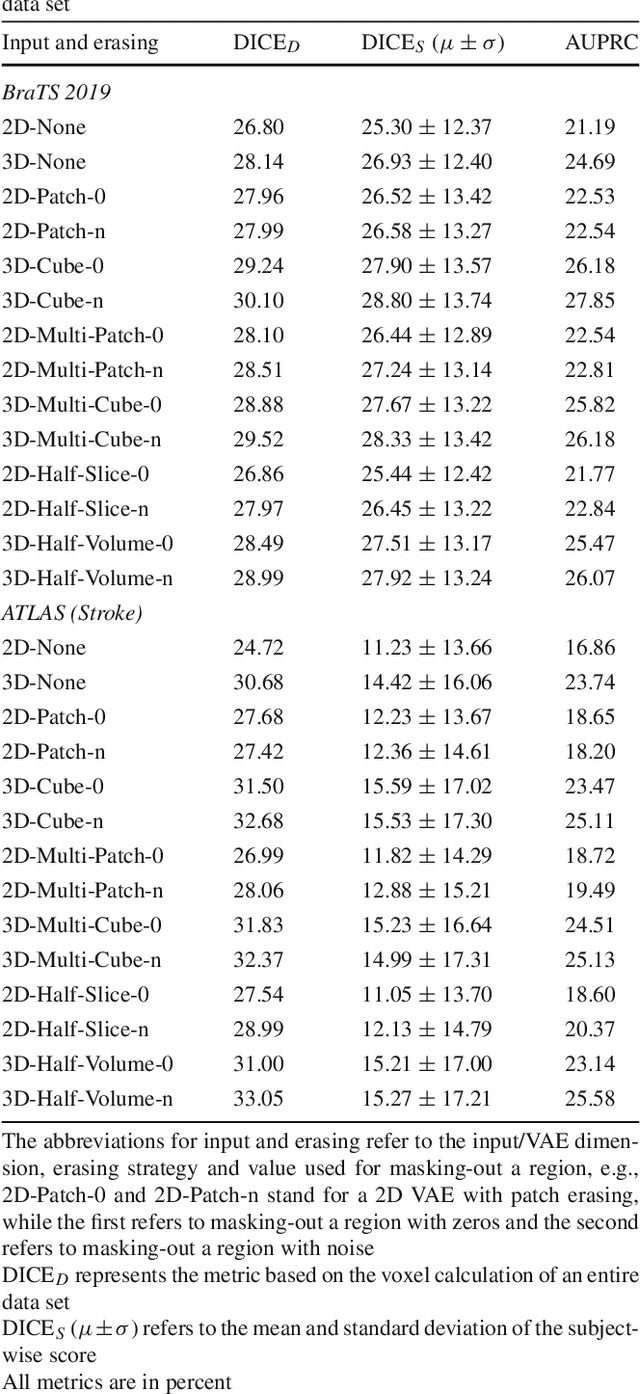
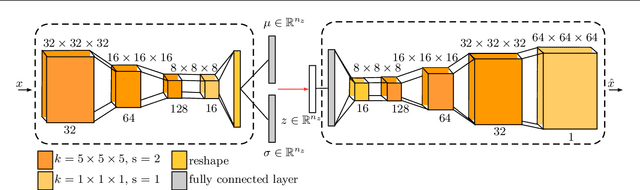
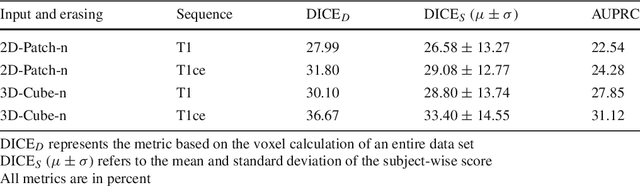
Purpose. Brain Magnetic Resonance Images (MRIs) are essential for the diagnosis of neurological diseases. Recently, deep learning methods for unsupervised anomaly detection (UAD) have been proposed for the analysis of brain MRI. These methods rely on healthy brain MRIs and eliminate the requirement of pixel-wise annotated data compared to supervised deep learning. While a wide range of methods for UAD have been proposed, these methods are mostly 2D and only learn from MRI slices, disregarding that brain lesions are inherently 3D and the spatial context of MRI volumes remains unexploited. Methods. We investigate whether using increased spatial context by using MRI volumes combined with spatial erasing leads to improved unsupervised anomaly segmentation performance compared to learning from slices. We evaluate and compare 2D variational autoencoder (VAE) to their 3D counterpart, propose 3D input erasing, and systemically study the impact of the data set size on the performance. Results. Using two publicly available segmentation data sets for evaluation, 3D VAE outperform their 2D counterpart, highlighting the advantage of volumetric context. Also, our 3D erasing methods allow for further performance improvements. Our best performing 3D VAE with input erasing leads to an average DICE score of 31.40% compared to 25.76% for the 2D VAE. Conclusions. We propose 3D deep learning methods for UAD in brain MRI combined with 3D erasing and demonstrate that 3D methods clearly outperform their 2D counterpart for anomaly segmentation. Also, our spatial erasing method allows for further performance improvements and reduces the requirement for large data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge