Zoltán Perkó
A Deep Learning approach for parametrized and time dependent Partial Differential Equations using Dimensionality Reduction and Neural ODEs
Feb 12, 2025Abstract:Partial Differential Equations (PDEs) are central to science and engineering. Since solving them is computationally expensive, a lot of effort has been put into approximating their solution operator via both traditional and recently increasingly Deep Learning (DL) techniques. A conclusive methodology capable of accounting both for (continuous) time and parameter dependency in such DL models however is still lacking. In this paper, we propose an autoregressive and data-driven method using the analogy with classical numerical solvers for time-dependent, parametric and (typically) nonlinear PDEs. We present how Dimensionality Reduction (DR) can be coupled with Neural Ordinary Differential Equations (NODEs) in order to learn the solution operator of arbitrary PDEs. The idea of our work is that it is possible to map the high-fidelity (i.e., high-dimensional) PDE solution space into a reduced (low-dimensional) space, which subsequently exhibits dynamics governed by a (latent) Ordinary Differential Equation (ODE). Solving this (easier) ODE in the reduced space allows avoiding solving the PDE in the high-dimensional solution space, thus decreasing the computational burden for repeated calculations for e.g., uncertainty quantification or design optimization purposes. The main outcome of this work is the importance of exploiting DR as opposed to the recent trend of building large and complex architectures: we show that by leveraging DR we can deliver not only more accurate predictions, but also a considerably lighter and faster DL model compared to existing methodologies.
Learning the Physics of Particle Transport via Transformers
Sep 08, 2021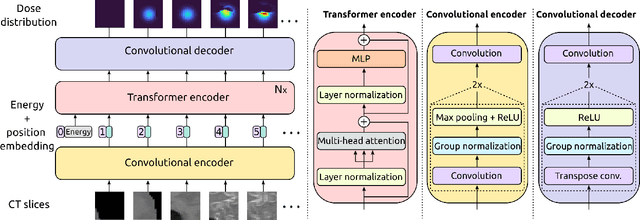
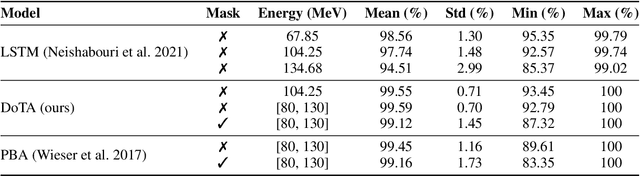
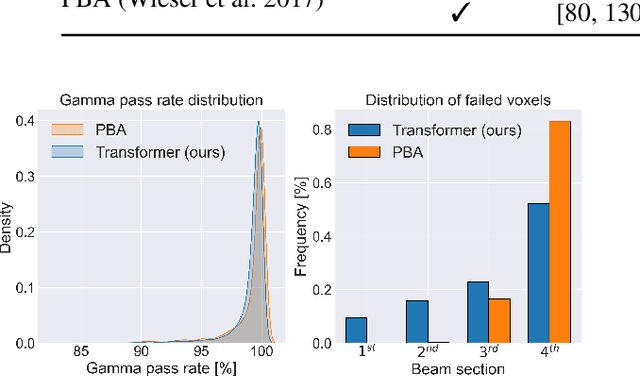
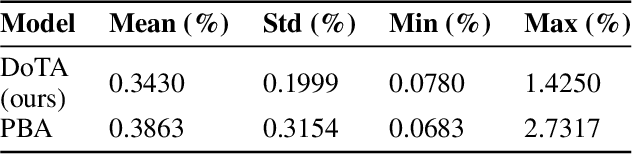
Abstract:Particle physics simulations are the cornerstone of nuclear engineering applications. Among them radiotherapy (RT) is crucial for society, with 50% of cancer patients receiving radiation treatments. For the most precise targeting of tumors, next generation RT treatments aim for real-time correction during radiation delivery, necessitating particle transport algorithms that yield precise dose distributions in sub-second times even in highly heterogeneous patient geometries. This is infeasible with currently available, purely physics based simulations. In this study, we present a data-driven dose calculation algorithm predicting the dose deposited by mono-energetic proton beams for arbitrary energies and patient geometries. Our approach frames particle transport as sequence modeling, where convolutional layers extract important spatial features into tokens and the transformer self-attention mechanism routes information between such tokens in the sequence and a beam energy token. We train our network and evaluate prediction accuracy using computationally expensive but accurate Monte Carlo (MC) simulations, considered the gold standard in particle physics. Our proposed model is 33 times faster than current clinical analytic pencil beam algorithms, improving upon their accuracy in the most heterogeneous and challenging geometries. With a relative error of 0.34% and very high gamma pass rate of 99.59% (1%, 3 mm), it also greatly outperforms the only published similar data-driven proton dose algorithm, even at a finer grid resolution. Offering MC precision 400 times faster, our model could overcome a major obstacle that has so far prohibited real-time adaptive proton treatments and significantly increase cancer treatment efficacy. Its potential to model physics interactions of other particles could also boost heavy ion treatment planning procedures limited by the speed of traditional methods.
Modeling biomedical breathing signals with convolutional deep probabilistic autoencoders
Oct 19, 2020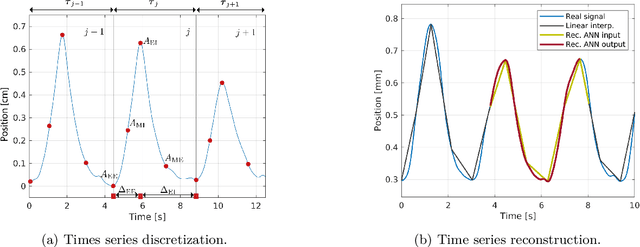
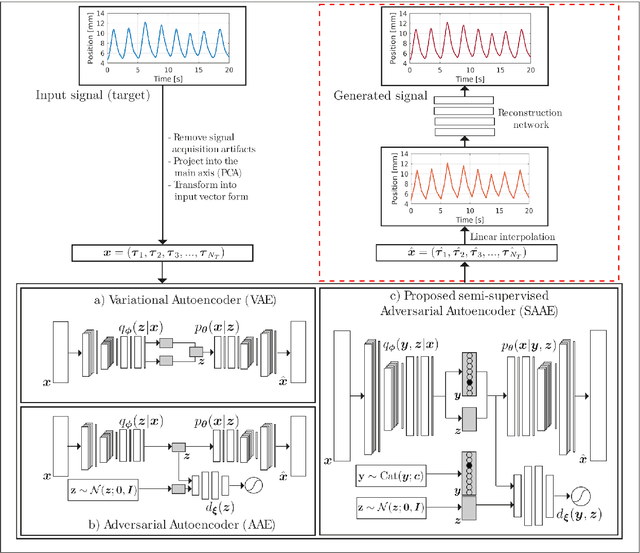
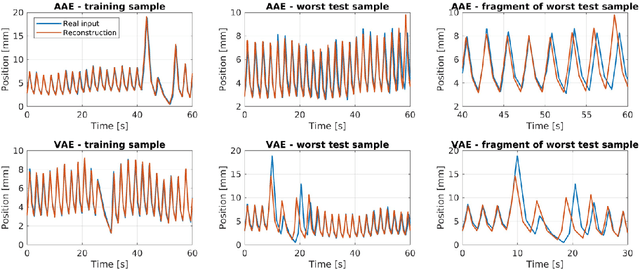
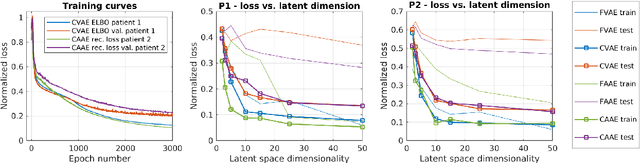
Abstract:One of the main problems with biomedical signals is the limited amount of patient-specific data and the significant amount of time needed to record a sufficient number of samples for diagnostic and treatment purposes. We explore the use of Variational Autoencoder (VAE) and Adversarial Autoencoder (AAE) algorithms based on one-dimensional convolutional neural networks in order to build generative models able to capture and represent the variability of a set of unlabeled quasi-periodic signals using as few as 10 parameters. Furthermore, we introduce a modified AAE architecture that allows simultaneous semi-supervised classification and generation of different types of signals. Our study is based on physical breathing signals, i.e. time series describing the position of chest markers, generally used to describe respiratory motion. The time series are discretized into a vector of periods, with each period containing 6 time and position values. These vectors can be transformed back into time series through an additional reconstruction neural network and allow to generate extended signals while simplifying the modeling task. The obtained models can be used to generate realistic breathing realizations from patient or population data and to classify new recordings. We show that by incorporating the labels from around 10-15\% of the dataset during training, the model can be guided to group data according to the patient it belongs to, or based on the presence of different types of breathing irregularities such as baseline shifts. Our specific motivation is to model breathing motion during radiotherapy lung cancer treatments, for which the developed model serves as an efficient tool to robustify plans against breathing uncertainties. However, the same methodology can in principle be applied to any other kind of quasi-periodic biomedical signal, representing a generically applicable tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge