Zhuoran Guo
NLICE: Synthetic Medical Record Generation for Effective Primary Healthcare Differential Diagnosis
Jan 24, 2024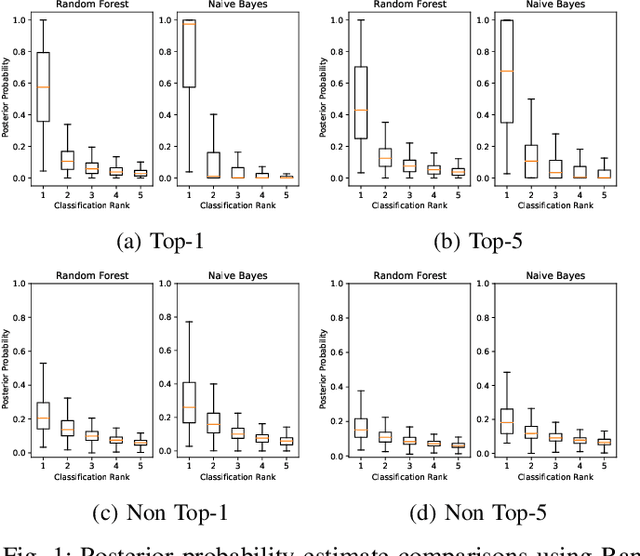
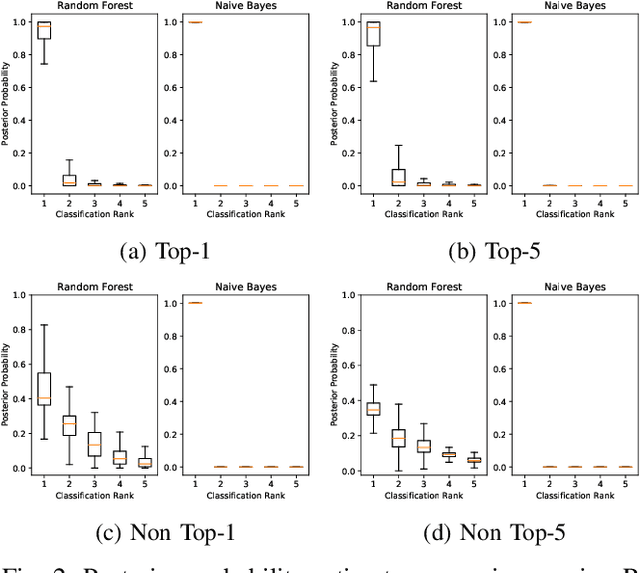
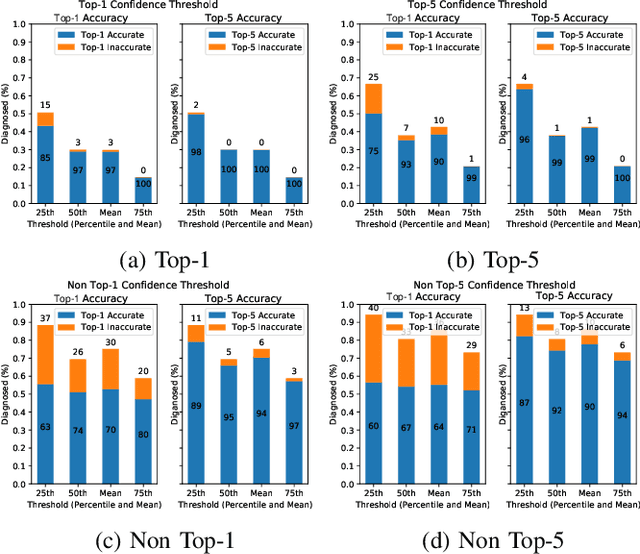
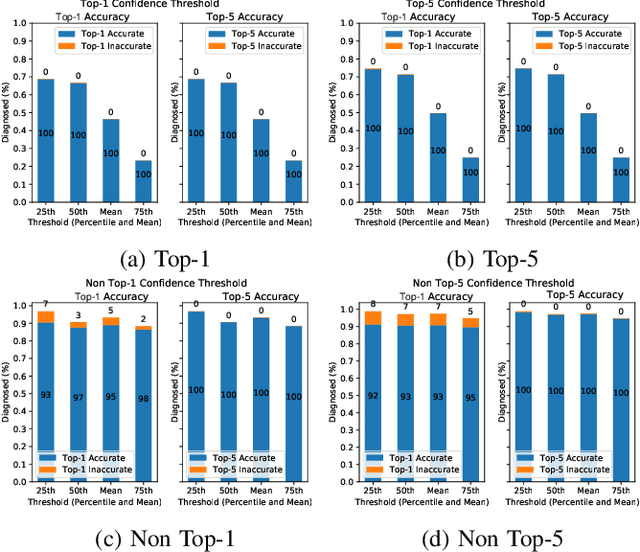
Abstract:This paper offers a systematic method for creating medical knowledge-grounded patient records for use in activities involving differential diagnosis. Additionally, an assessment of machine learning models that can differentiate between various conditions based on given symptoms is also provided. We use a public disease-symptom data source called SymCat in combination with Synthea to construct the patients records. In order to increase the expressive nature of the synthetic data, we use a medically-standardized symptom modeling method called NLICE to augment the synthetic data with additional contextual information for each condition. In addition, Naive Bayes and Random Forest models are evaluated and compared on the synthetic data. The paper shows how to successfully construct SymCat-based and NLICE-based datasets. We also show results for the effectiveness of using the datasets to train predictive disease models. The SymCat-based dataset is able to train a Naive Bayes and Random Forest model yielding a 58.8% and 57.1% Top-1 accuracy score, respectively. In contrast, the NLICE-based dataset improves the results, with a Top-1 accuracy of 82.0% and Top-5 accuracy values of more than 90% for both models. Our proposed data generation approach solves a major barrier to the application of artificial intelligence methods in the healthcare domain. Our novel NLICE symptom modeling approach addresses the incomplete and insufficient information problem in the current binary symptom representation approach. The NLICE code is open sourced at https://github.com/guozhuoran918/NLICE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge