Zhenyu Meng
Persistent spectral based machine learning (PerSpect ML) for drug design
Feb 03, 2020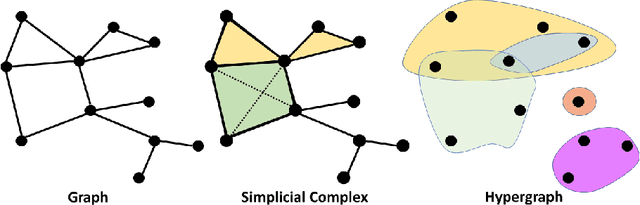
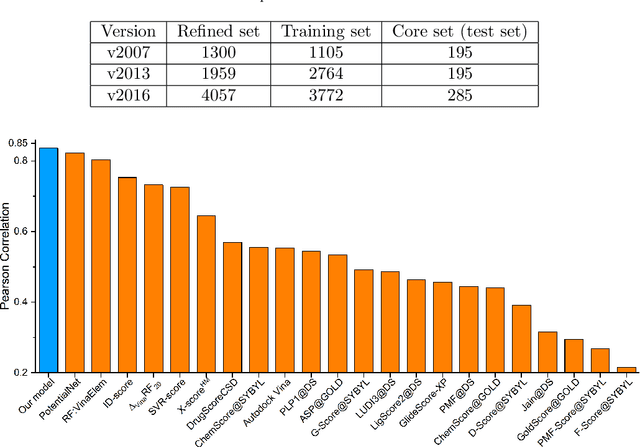
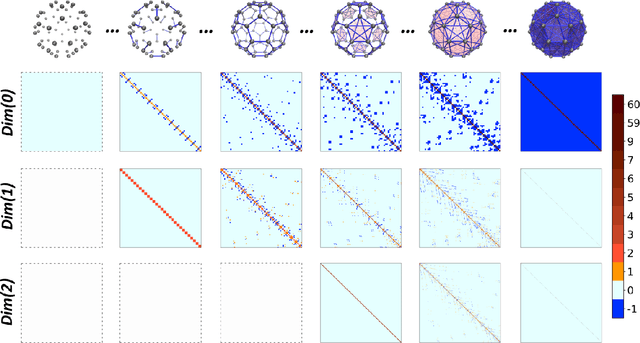

Abstract:In this paper, we propose persistent spectral based machine learning (PerSpect ML) models for drug design. Persistent spectral models, including persistent spectral graph, persistent spectral simplicial complex and persistent spectral hypergraph, are proposed based on spectral graph theory, spectral simplicial complex theory and spectral hypergraph theory, respectively. Different from all previous spectral models, a filtration process, as proposed in persistent homology, is introduced to generate multiscale spectral models. More specifically, from the filtration process, a series of nested topological representations, i,e., graphs, simplicial complexes, and hypergraphs, can be systematically generated and their spectral information can be obtained. Persistent spectral variables are defined as the function of spectral variables over the filtration value. Mathematically, persistent multiplicity (of zero eigenvalues) is exactly the persistent Betti number (or Betti curve). We consider 11 persistent spectral variables and use them as the feature for machine learning models in protein-ligand binding affinity prediction. We systematically test our models on three most commonly-used databases, including PDBbind-2007, PDBbind-2013 and PDBbind-2016. Our results, for all these databases, are better than all existing models, as far as we know. This demonstrates the great power of our PerSpect ML in molecular data analysis and drug design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge