Yunbi Nam
Recent advances in deep learning and language models for studying the microbiome
Sep 15, 2024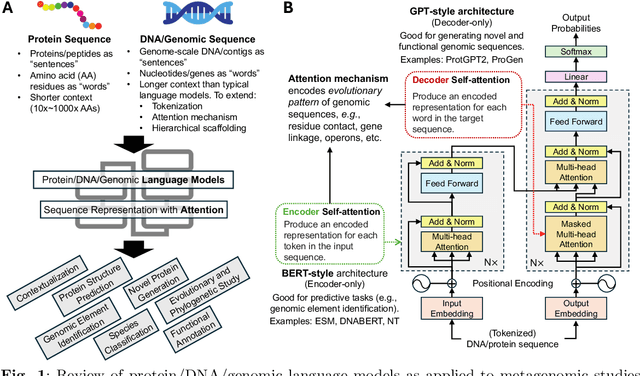

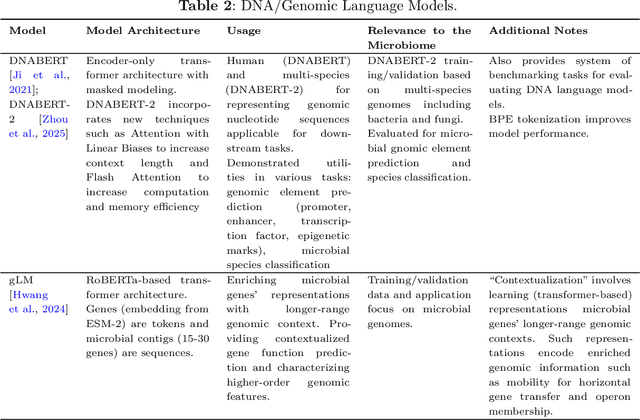

Abstract:Recent advancements in deep learning, particularly large language models (LLMs), made a significant impact on how researchers study microbiome and metagenomics data. Microbial protein and genomic sequences, like natural languages, form a language of life, enabling the adoption of LLMs to extract useful insights from complex microbial ecologies. In this paper, we review applications of deep learning and language models in analyzing microbiome and metagenomics data. We focus on problem formulations, necessary datasets, and the integration of language modeling techniques. We provide an extensive overview of protein/genomic language modeling and their contributions to microbiome studies. We also discuss applications such as novel viromics language modeling, biosynthetic gene cluster prediction, and knowledge integration for metagenomics studies.
Random Forest Variable Importance-based Selection Algorithm in Class Imbalance Problem
Dec 17, 2023Abstract:Random Forest is a machine learning method that offers many advantages, including the ability to easily measure variable importance. Class balancing technique is a well-known solution to deal with class imbalance problem. However, it has not been actively studied on RF variable importance. In this paper, we study the effect of class balancing on RF variable importance. Our simulation results show that over-sampling is effective in correctly measuring variable importance in class imbalanced situations with small sample size, while under-sampling fails to differentiate important and non-informative variables. We then propose a variable selection algorithm that utilizes RF variable importance and its confidence interval. Through an experimental study using many real and artificial datasets, we demonstrate that our proposed algorithm efficiently selects an optimal feature set, leading to improved prediction performance in class imbalance problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge