Youngsun Kong
Autoencoder-Based Denoising of Muscle Artifacts in ECG to Preserve Skin Nerve Activity (SKNA) for Cognitive Stress Detection
Sep 08, 2025



Abstract:The sympathetic nervous system (SNS) plays a central role in regulating the body's responses to stress and maintaining physiological stability. Its dysregulation is associated with a wide range of conditions, from cardiovascular disease to anxiety disorders. Skin nerve activity (SKNA) extracted from high-frequency electrocardiogram (ECG) recordings provides a noninvasive window into SNS dynamics, but its measurement is highly susceptible to electromyographic (EMG) contamination. Traditional preprocessing based on bandpass filtering within a fixed range (e.g., 500--1000 Hz) is susceptible to overlapping EMG and SKNA spectral components, especially during sustained muscle activity. We present a denoising approach using a lightweight one-dimensional convolutional autoencoder with a long short-term memory (LSTM) bottleneck to reconstruct clean SKNA from EMG-contaminated recordings. Using clean ECG-derived SKNA data from cognitive stress experiments and EMG noise from chaotic muscle stimulation recordings, we simulated contamination at realistic noise levels (--4 dB, --8 dB signal-to-noise ratio) and trained the model in the leave-one-subject-out cross-validation framework. The method improved signal-to-noise ratio by up to 9.65 dB, increased cross correlation with clean SKNA from 0.40 to 0.72, and restored burst-based SKNA features to near-clean discriminability (AUROC $\geq$ 0.96). Classification of baseline versus sympathetic stimulation (cognitive stress) conditions reached accuracies of 91--98\% across severe noise levels, comparable to clean data. These results demonstrate that deep learning--based reconstruction can preserve physiologically relevant sympathetic bursts during substantial EMG interference, enabling more robust SKNA monitoring in naturalistic, movement-rich environments.
A Novel Approach to Characterize Dynamics of ECG-Derived Skin Nerve Activity via Time-Varying Spectral Analysis
Nov 13, 2024Abstract:Assessment of the sympathetic nervous system (SNS) is one of the major approaches for studying affective states. Skin nerve activity (SKNA) derived from high-frequency components of electrocardiogram (ECG) signals has been a promising surrogate for assessing the SNS. However, current SKNA analysis tools have shown high variability across study protocols and experiments. Hence, we propose a time-varying spectral approach based on SKNA to assess the SNS with higher sensitivity and reliability. We collected ECG signals at a sampling frequency of 10 KHz from sixteen subjects who underwent various SNS stimulations. Our spectral analysis revealed that frequency bands between 150 - 1,000 Hz showed significant increases in power during SNS stimulations. Using this information, we developed a time-varying index of sympathetic function measurement based on SKNA, termed, Time-Varying Skin Nerve Activity (TVSKNA). TVSKNA is calculated in three steps: time-frequency decomposition, reconstruction using selected frequency bands, and smoothing. TVSKNA indices exhibited generally higher Youden's J, balanced accuracy, and area under the receiver operating characteristic curve, indicating higher sensitivity. The coefficient of variance was lower with TVSKNA indices for most SNS tasks. TVSKNA can serve as a highly sensitive and reliable marker of quantitative assessment of sympathetic function, especially during emotion and stress.
Towards Continuous Skin Sympathetic Nerve Activity Monitoring: Removing Muscle Noise
Oct 26, 2024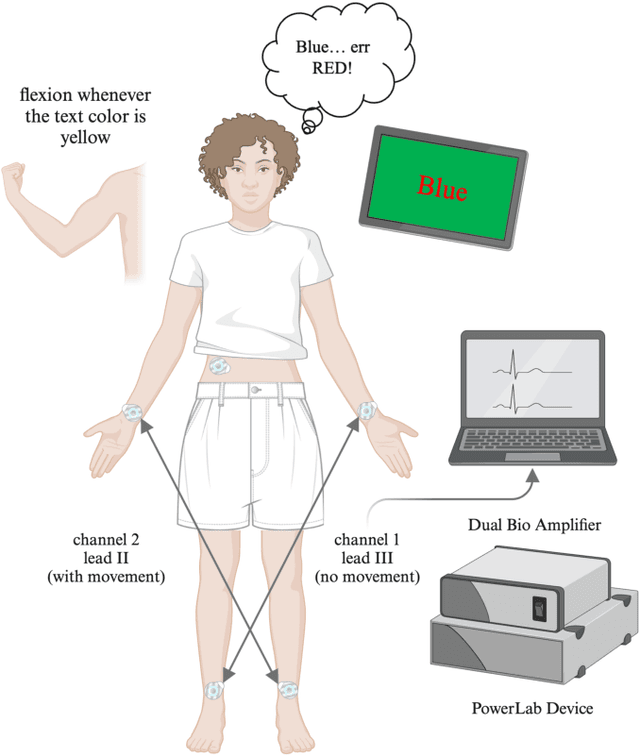



Abstract:Continuous monitoring of non-invasive skin sympathetic nerve activity (SKNA) holds promise for understanding the sympathetic nervous system (SNS) dynamics in various physiological and pathological conditions. However, muscle noise artifacts present a challenge in accurate SKNA analysis, particularly in real-life scenarios. This study proposes a deep convolutional neural network (CNN) approach to detect and remove muscle noise from SKNA recordings obtained via ECG electrodes. Twelve healthy participants underwent controlled experimental protocols involving cognitive stress induction and voluntary muscle movements, while collecting SKNA data. Power spectral analysis revealed significant muscle noise interference within the SKNA frequency band (500-1000 Hz). A 2D CNN model was trained on the spectrograms of the data segments to classify them into baseline, stress-induced SKNA, and muscle noise-contaminated periods, achieving an average accuracy of 89.85% across all subjects. Our findings underscore the importance of addressing muscle noise for accurate SKNA monitoring, advancing towards wearable SKNA sensors for real-world applications.
A Preliminary Study on Automatic Motion Artifacts Detection in Electrodermal Activity Data Using Machine Learning
Jul 16, 2021



Abstract:The electrodermal activity (EDA) signal is a sensitive and non-invasive surrogate measure of sympathetic function. Use of EDA has increased in popularity in recent years for such applications as emotion and stress recognition; assessment of pain, fatigue, and sleepiness; diagnosis of depression and epilepsy; and other uses. Recently, there have been several studies using ambulatory EDA recordings, which are often quite useful for analysis of many physiological conditions. Because ambulatory monitoring uses wearable devices, EDA signals are often affected by noise and motion artifacts. An automated noise and motion artifact detection algorithm is therefore of utmost importance for accurate analysis and evaluation of EDA signals. In this paper, we present machine learning-based algorithms for motion artifact detection in EDA signals. With ten subjects, we collected two simultaneous EDA signals from the right and left hands, while instructing the subjects to move only the right hand. Using these data, we proposed a cross-correlation-based approach for non-biased labeling of EDA data segments. A set of statistical, spectral and model-based features were calculated which were then subjected to a feature selection algorithm. Finally, we trained and validated several machine learning methods using a leave-one-subject-out approach. The classification accuracy of the developed model was 83.85% with a standard deviation of 4.91%, which was better than a recent standard method that we considered for comparison to our algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge