Yeojin Kim
PINNet: a deep neural network with pathway prior knowledge for Alzheimer's disease
Nov 27, 2022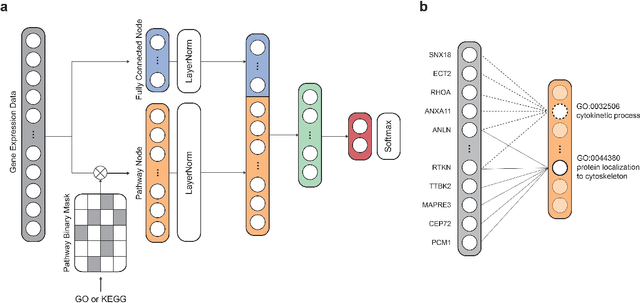

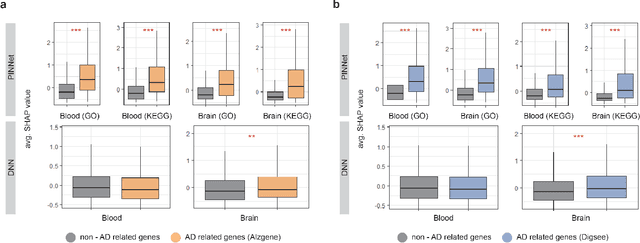
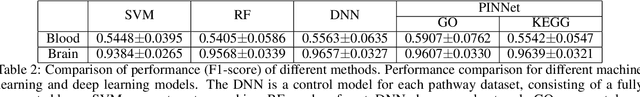
Abstract:Identification of Alzheimer's Disease (AD)-related transcriptomic signatures from blood is important for early diagnosis of the disease. Deep learning techniques are potent classifiers for AD diagnosis, but most have been unable to identify biomarkers because of their lack of interpretability. To address these challenges, we propose a pathway information-based neural network (PINNet) to predict AD patients and analyze blood and brain transcriptomic signatures using an interpretable deep learning model. PINNet is a deep neural network (DNN) model with pathway prior knowledge from either the Gene Ontology or Kyoto Encyclopedia of Genes and Genomes databases. Then, a backpropagation-based model interpretation method was applied to reveal essential pathways and genes for predicting AD. We compared the performance of PINNet with a DNN model without a pathway. Performances of PINNet outperformed or were similar to those of DNN without a pathway using blood and brain gene expressions, respectively. Moreover, PINNet considers more AD-related genes as essential features than DNN without a pathway in the learning process. Pathway analysis of protein-protein interaction modules of highly contributed genes showed that AD-related genes in blood were enriched with cell migration, PI3K-Akt, MAPK signaling, and apoptosis in blood. The pathways enriched in the brain module included cell migration, PI3K-Akt, MAPK signaling, apoptosis, protein ubiquitination, and t-cell activation. Collectively, with prior knowledge about pathways, PINNet reveals essential pathways related to AD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge