Yanhua Gao
Department of Ultrasound, Shaanxi Provincial People's Hospital, Xi'an, China
Preparing data for pathological artificial intelligence with clinical-grade performance
May 22, 2022



Abstract:[Purpose] The pathology is decisive for disease diagnosis, but relies heavily on the experienced pathologists. Recently, pathological artificial intelligence (PAI) is thought to improve diagnostic accuracy and efficiency. However, the high performance of PAI based on deep learning in the laboratory generally cannot be reproduced in the clinic. [Methods] Because the data preparation is important for PAI, the paper has reviewed PAI-related studies in the PubMed database published from January 2017 to February 2022, and 118 studies were included. The in-depth analysis of methods for preparing data is performed, including obtaining slides of pathological tissue, cleaning, screening, and then digitizing. Expert review, image annotation, dataset division for model training and validation are also discussed. We further discuss the reasons why the high performance of PAI is not reproducible in the clinical practices and show some effective ways to improve clinical performances of PAI. [Results] The robustness of PAI depend on randomized collection of representative disease slides, including rigorous quality control and screening, correction of digital discrepancies, reasonable annotation, and the amount of data. The digital pathology is fundamental of clinical-grade PAI, and the techniques of data standardization and weakly supervised learning methods based on whole slide image (WSI) are effective ways to overcome obstacles of performance reproduction. [Conclusion] The representative data, the amount of labeling and consistency from multi-centers is the key to performance reproduction. The digital pathology for clinical diagnosis, data standardization and technique of WSI-based weakly supervised learning hopefully build clinical-grade PAI. Keywords: pathological artificial intelligence; data preparation; clinical-grade; deep learning
Multi-scale super-resolution generation of low-resolution scanned pathological images
May 15, 2021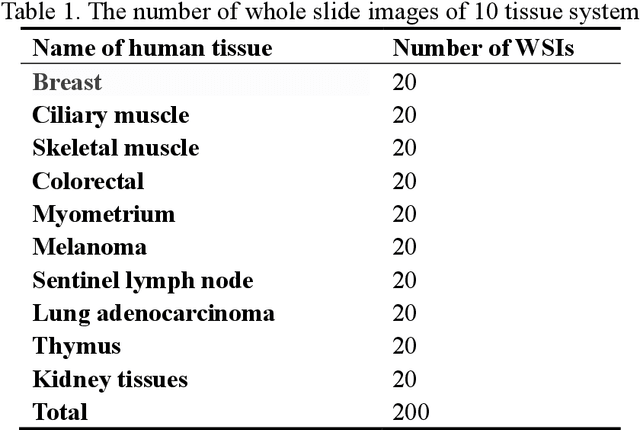

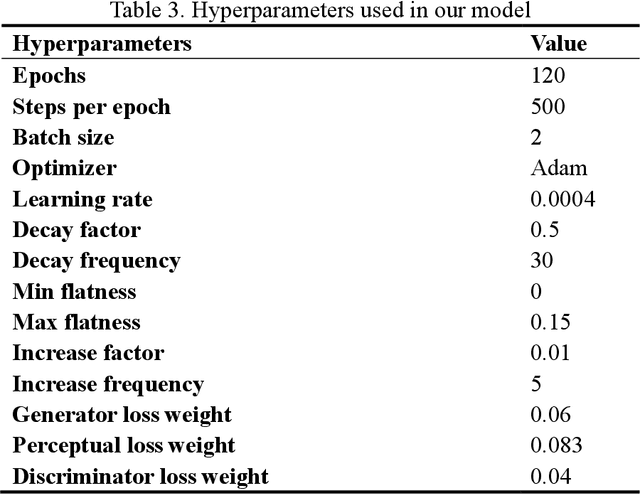
Abstract:Digital pathology slide is easy to store and manage, convenient to browse and transmit. However, because of the high-resolution scan for example 40 times magnification(40X) during the digitization, the file size of each whole slide image exceeds 1Gigabyte, which eventually leads to huge storage capacity and very slow network transmission. We design a strategy to scan slides with low resolution (5X) and a super-resolution method is proposed to restore the image details when in diagnosis. The method is based on a multi-scale generative adversarial network, which sequentially generate three high-resolution images such as 10X, 20X and 40X. The perceived loss, generator loss of the generated images and real images are compared on three image resolutions, and a discriminator is used to evaluate the difference of highest-resolution generated image and real image. A dataset consisting of 100,000 pathological images from 10 types of human tissues is performed for training and testing the network. The generated images have high peak-signal-to-noise-ratio (PSNR) and structural-similarity-index (SSIM). The PSNR of 10X to 40X image are 24.16, 22.27 and 20.44, and the SSIM are 0.845, 0.680 and 0.512, which are better than other super-resolution networks such as DBPN, ESPCN, RDN, EDSR and MDSR. Moreover, visual inspections show that the generated high-resolution images by our network have enough details for diagnosis, good color reproduction and close to real images, while other five networks are severely blurred, local deformation or miss important details. Moreover, no significant differences can be found on pathological diagnosis based on the generated and real images. The proposed multi-scale network can generate good high-resolution pathological images, and will provide a low-cost storage (about 15MB/image on 5X), faster image sharing method for digital pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge