Xueyuan Zhao
Source Feature Compression for Object Classification in Vision-Based Underwater Robotics
Dec 28, 2021


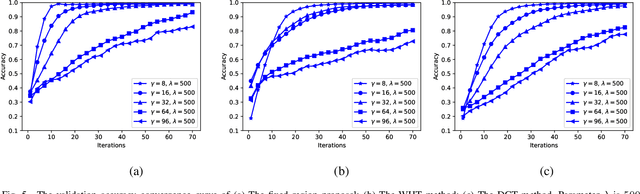
Abstract:New efficient source feature compression solutions are proposed based on a two-stage Walsh-Hadamard Transform (WHT) for Convolutional Neural Network (CNN)-based object classification in underwater robotics. The object images are firstly transformed by WHT following a two-stage process. The transform-domain tensors have large values concentrated in the upper left corner of the matrices in the RGB channels. By observing this property, the transform-domain matrix is partitioned into inner and outer regions. Consequently, two novel partitioning methods are proposed in this work: (i) fixing the size of inner and outer regions; and (ii) adjusting the size of inner and outer regions adaptively per image. The proposals are evaluated with an underwater object dataset captured from the Raritan River in New Jersey, USA. It is demonstrated and verified that the proposals reduce the training time effectively for learning-based underwater object classification task and increase the accuracy compared with the competing methods. The object classification is an essential part of a vision-based underwater robot that can sense the environment and navigate autonomously. Therefore, the proposed method is well-suited for efficient computer vision-based tasks in underwater robotics applications.
Configuration Learning in Underwater Optical Links
Aug 03, 2020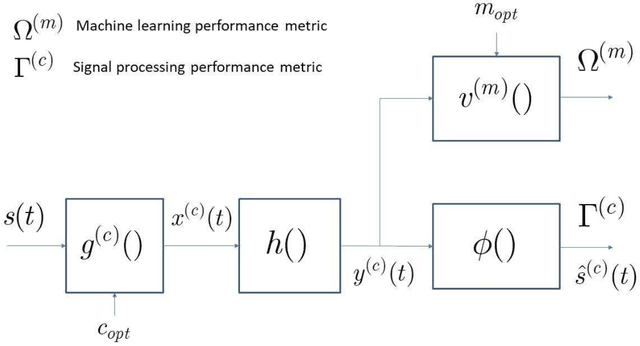
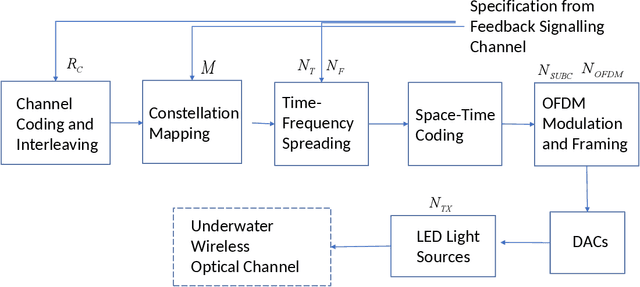
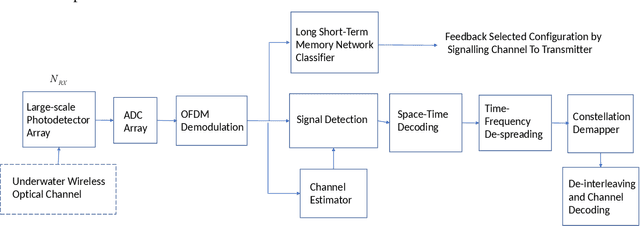
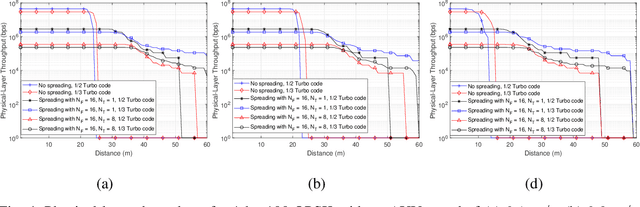
Abstract:A new research problem named configuration learning is described in this work. A novel algorithm is proposed to address the configuration learning problem. The configuration learning problem is defined to be the optimization of the Machine Learning (ML) classifier to maximize the ML performance metric optimizing the transmitter configuration in the signal processing/communication systems. Specifically, this configuration learning problem is investigated in an underwater optical communication system with signal processing performance metric of the physical-layer communication throughput. A novel algorithm is proposed to perform the configuration learning by alternating optimization of key design parameters and switching between several Recurrent Neural Network (RNN) classifiers dependant on the learning objective. The proposed ML algorithm is validated with the datasets of an underwater optical communication system and is compared with competing ML algorithms. Performance results indicate that the proposal outperforms the competing algorithms for binary and multi-class configuration learning in underwater optical communication datasets. The proposed configuration learning framework can be further investigated and applied to a broad range of topics in signal processing and communications.
Transform-Domain Classification of Human Cells based on DNA Methylation Datasets
Dec 31, 2019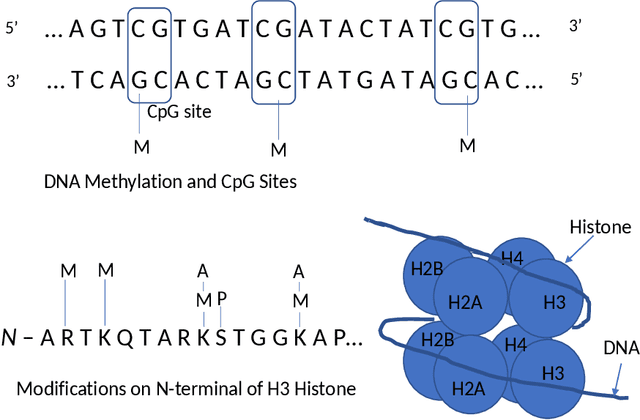
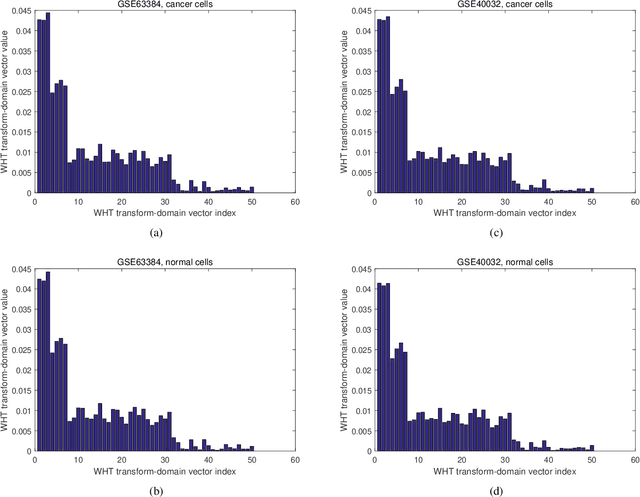


Abstract:A novel method to classify human cells is presented in this work based on the transform-domain method on DNA methylation data. DNA methylation profile variations are observed in human cells with the progression of disease stages, and the proposal is based on this DNA methylation variation to classify normal and disease cells including cancer cells. The cancer cell types investigated in this work cover hepatocellular (sample size n = 40), colorectal (n = 44), lung (n = 70) and endometrial (n = 87) cancer cells. A new pipeline is proposed integrating the DNA methylation intensity measurements on all the CpG islands by the transformation of Walsh-Hadamard Transform (WHT). The study reveals the three-step properties of the DNA methylation transform-domain data and the step values of association with the cell status. Further assessments have been carried out on the proposed machine learning pipeline to perform classification of the normal and cancer tissue cells. A number of machine learning classifiers are compared for whole sequence and WHT sequence classification based on public Whole-Genome Bisulfite Sequencing (WGBS) DNA methylation datasets. The WHT-based method can speed up the computation time by more than one order of magnitude compared with whole original sequence classification, while maintaining comparable classification accuracy by the selected machine learning classifiers. The proposed method has broad applications in expedited disease and normal human cell classifications by the epigenome and genome datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge