Vishal Srivastava
Incorporating Total Variation Regularization in the design of an intelligent Query by Humming system
Feb 09, 2023



Abstract:A Query-By-Humming (QBH) system constitutes a particular case of music information retrieval where the input is a user-hummed melody and the output is the original song which contains that melody. A typical QBH system consists of melody extraction and candidate melody retrieval. For melody extraction, accurate note transcription is the key enabling technology. However, current transcription methods are unable to definitively capture the melody and address inaccuracies in user-hummed queries. In this paper, we incorporate Total Variation Regularization (TVR) to denoise queries. This approach accounts for user error in humming without loss of meaningful data and reliably captures the underlying melody. For candidate melody retrieval, we employ a deep learning approach to time series classification using a Fully Convolutional Neural Network. The trained network classifies the incoming query as belonging to one of the target songs. For our experiments, we use Roger Jang's MIR-QBSH dataset which is the standard MIREX dataset. We demonstrate that inclusion of TVR denoised queries in the training set enhances the overall accuracy of the system to 93% which is higher than other state-of-the-art QBH systems.
Self-attention based BiLSTM-CNN classifier for the prediction of ischemic and non-ischemic cardiomyopathy
Jul 29, 2019
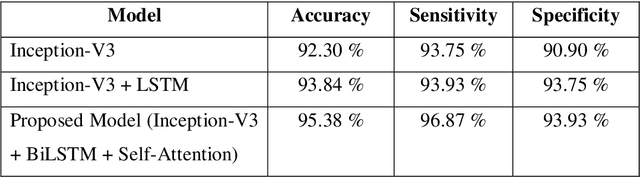


Abstract:Heart Failure is a major component of healthcare expenditure and a leading cause of mortality worldwide. Despite higher inter-rater variability, endomyocardial biopsy (EMB) is still regarded as the standard technique, used to identify the cause (e.g. ischemic or non-ischemic cardiomyopathy, coronary artery disease, myocardial infarction etc.) of unexplained heart failure. In this paper, we focus on identifying cardiomyopathy as ischemic or non-ischemic. For this, we propose and implement a new unified architecture comprising CNN (inception-V3 model) and bidirectional LSTM (BiLSTM) with self-attention mechanism to predict the ischemic or non-ischemic to classify cardiomyopathy using histopathological images. The proposed model is based on self-attention that implicitly focuses on the information outputted from the hidden layers of BiLSTM. Through our results we demonstrate that this framework carries a high learning capacity and is able to improve the classification performance.
Deep learning enabled multi-wavelength spatial coherence microscope for the classification of malaria-infected stages with limited labelled data size
Mar 14, 2019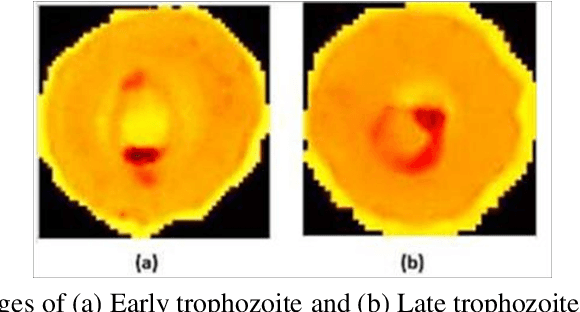
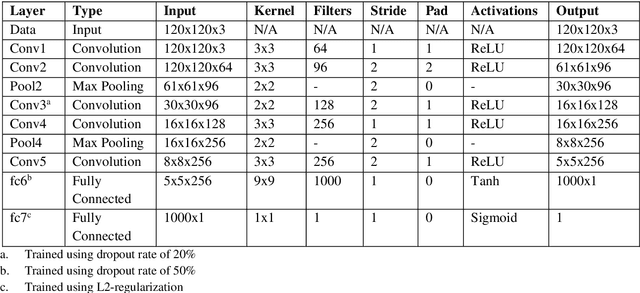
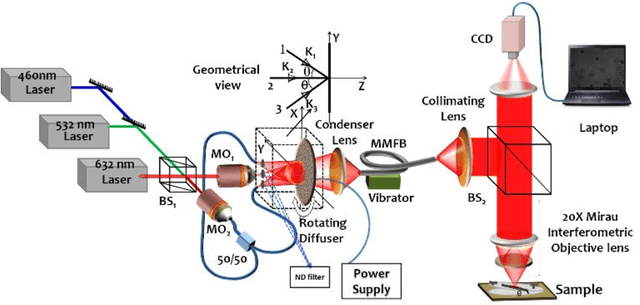
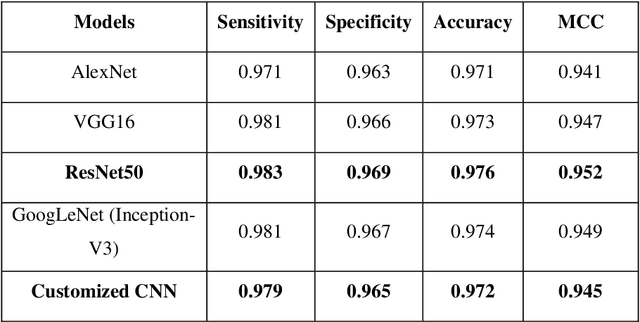
Abstract:Malaria is a life-threatening mosquito-borne blood disease, hence early detection is very crucial for health. The conventional method for the detection is a microscopic examination of Giemsa-stained blood smears, which needs a highly trained skilled technician. Automated classifications of different stages of malaria still a challenging task, especially having poor sensitivity in detecting the early trophozoite and late trophozoite or schizont stage with limited labelled datasize. The study aims to develop a fast, robust and fully automated system for the classification of different stages of malaria with limited data size by using the pre-trained convolutional neural networks (CNNs) as a classifier and multi-wavelength to increase the sample size. We also compare our customized CNN with other well-known CNNs and shows that our network have a comparable performance with less computational time. We believe that our proposed method can be applied to other limited labelled biological datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge