Vasundhara Dehiya
Prior Knowledge based mutation prioritization towards causal variant finding in rare disease
Oct 10, 2017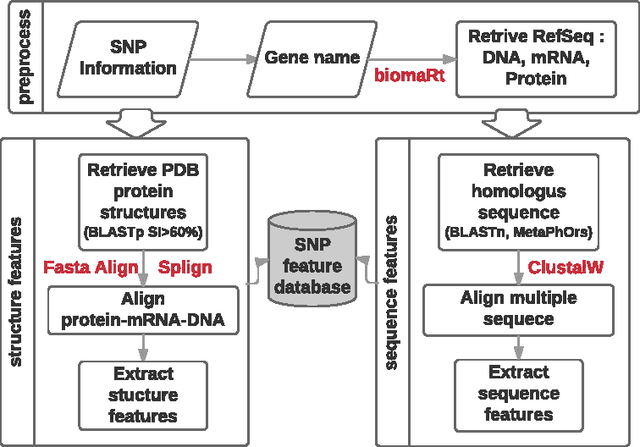
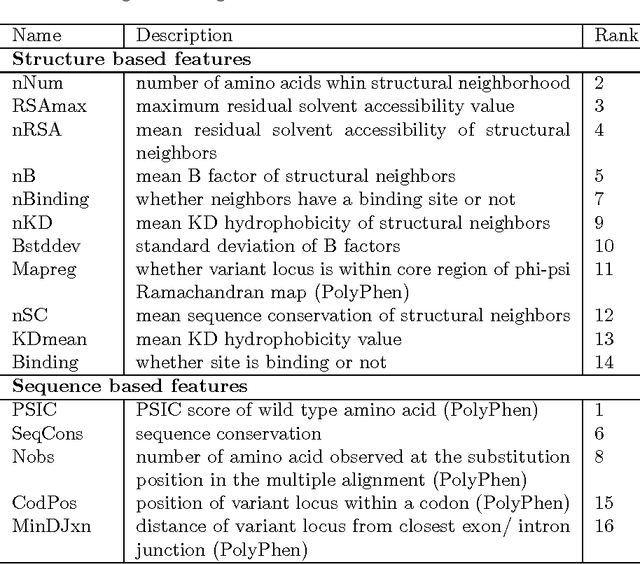
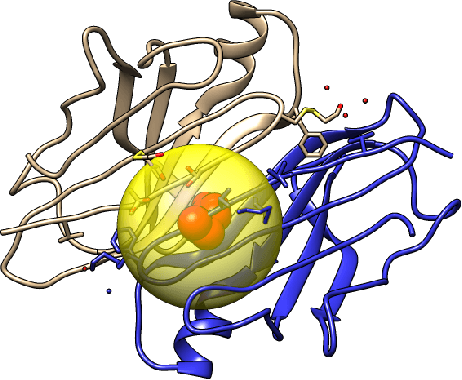
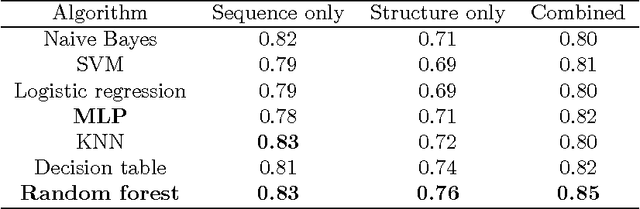
Abstract:How do we determine the mutational effects in exome sequencing data with little or no statistical evidence? Can protein structural information fill in the gap of not having enough statistical evidence? In this work, we answer the two questions with the goal towards determining pathogenic effects of rare variants in rare disease. We take the approach of determining the importance of point mutation loci focusing on protein structure features. The proposed structure-based features contain information about geometric, physicochemical, and functional information of mutation loci and those of structural neighbors of the loci. The performance of the structure-based features trained on 80\% of HumDiv and tested on 20\% of HumDiv and on ClinVar datasets showed high levels of discernibility in the mutation's pathogenic or benign effects: F score of 0.71 and 0.68 respectively using multi-layer perceptron. Combining structure- and sequence-based feature further improve the accuracy: F score of 0.86 (HumDiv) and 0.75 (ClinVar). Also, careful examination of the rare variants in rare diseases cases showed that structure-based features are important in discerning importance of variant loci.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge