Usman W. Roshan
A fully 3D multi-path convolutional neural network with feature fusion and feature weighting for automatic lesion identification in brain MRI images
Jul 17, 2019


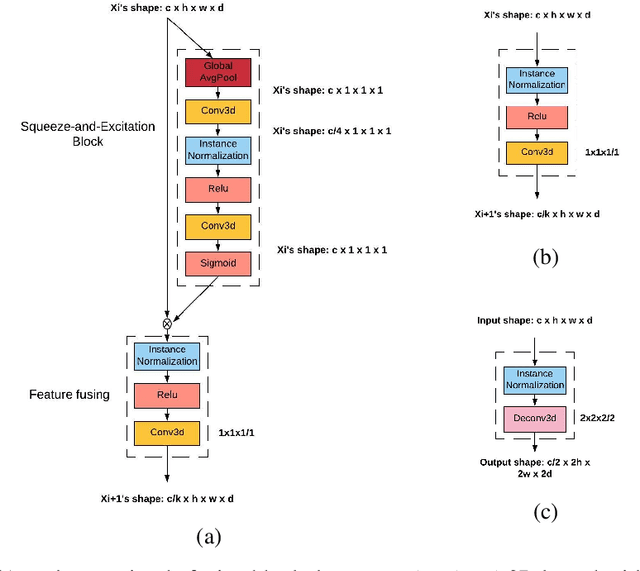
Abstract:Brain MRI images consist of multiple 2D images stacked at consecutive spatial intervals to form a 3D structure. Thus it seems natural to use a convolutional neural network with 3D convolutional kernels that would automatically also account for spatial dependence between the slices. However, 3D models remain a challenge in practice due to overfitting caused by insufficient training data. For example in a 2D model we typically have 150-300 slices per patient per plane of orientation whereas in a 3D setting this gets reduced to just one point. Here we propose a fully 3D multi-path convolutional network with custom designed components to better utilize features from multiple modalities. In particular our multi-path model has independent encoders for different modalities containing residual convolutional blocks, weighted multi-path feature fusion from different modalities, and weighted fusion modules to combine encoder and decoder features. We provide intuitive reasoning for different components along with empirical evidence to show that they work. Compared to existing 3D CNNs like DeepMedic, 3D U-Net, and AnatomyNet, our networks achieves the highest statistically significant cross-validation accuracy of 60.5% on the large ATLAS benchmark of 220 patients. We also test our model on multi-modal images from the Kessler Foundation and Medical College Wisconsin and achieve a statistically significant cross-validation accuracy of 65%, significantly outperforming the multi-modal 3D U-Net and DeepMedic. Overall our model offers a principled, extensible multi-path approach that outperforms multi-channel alternatives and achieves high Dice accuracies on existing benchmarks.
A multi-path 2.5 dimensional convolutional neural network system for segmenting stroke lesions in brain MRI images
May 26, 2019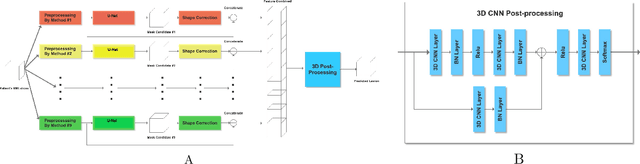



Abstract:Automatic identification of brain lesions from magnetic resonance imaging (MRI) scans of stroke survivors would be a useful aid in patient diagnosis and treatment planning. We propose a multi-modal multi-path convolutional neural network system for automating stroke lesion segmentation. Our system has nine end-to-end UNets that take as input 2-dimensional (2D) slices and examines all three planes with three different normalizations. Outputs from these nine total paths are concatenated into a 3D volume that is then passed to a 3D convolutional neural network to output a final lesion mask. We trained and tested our method on datasets from three sources: Medical College of Wisconsin (MCW), Kessler Foundation (KF), and the publicly available Anatomical Tracings of Lesions After Stroke (ATLAS) dataset. Cross-study validation results (with independent training and validation datasets) were obtained to compare with previous methods based on naive Bayes, random forests, and three recently published convolutional neural networks. Model performance was quantified in terms of the Dice coefficient. Training on the KF and MCW images and testing on the ATLAS images yielded a mean Dice coefficient of 0.54. This was reliably better than the next best previous model, UNet, at 0.47. Reversing the train and test datasets yields a mean Dice of 0.47 on KF and MCW images, whereas the next best UNet reaches 0.45. With all three datasets combined, the current system compared to previous methods also attained a reliably higher cross-validation accuracy. It also achieved high Dice values for many smaller lesions that existing methods have difficulty identifying. Overall, our system is a clear improvement over previous methods for automating stroke lesion segmentation, bringing us an important step closer to the inter-rater accuracy level of human experts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge