Ujwala Kiran Chaudhari
HistoSmith: Single-Stage Histology Image-Label Generation via Conditional Latent Diffusion for Enhanced Cell Segmentation and Classification
Feb 12, 2025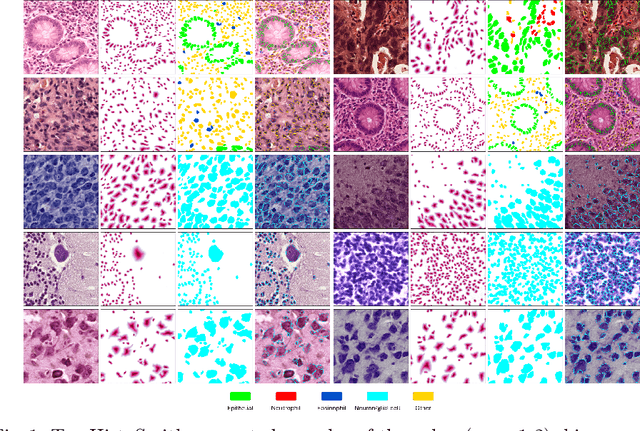
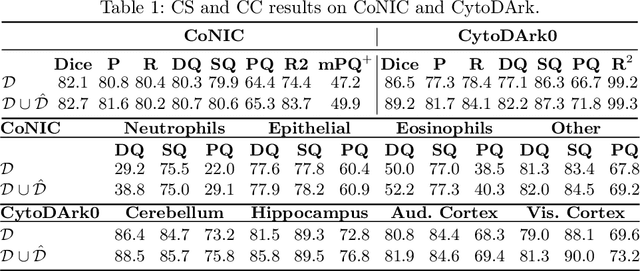
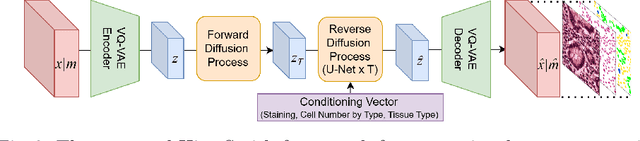
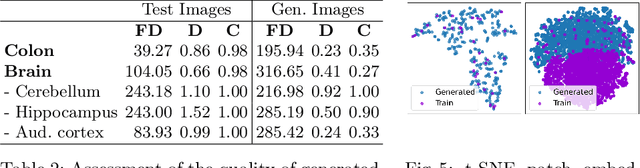
Abstract:Precise segmentation and classification of cell instances are vital for analyzing the tissue microenvironment in histology images, supporting medical diagnosis, prognosis, treatment planning, and studies of brain cytoarchitecture. However, the creation of high-quality annotated datasets for training remains a major challenge. This study introduces a novel single-stage approach (HistoSmith) for generating image-label pairs to augment histology datasets. Unlike state-of-the-art methods that utilize diffusion models with separate components for label and image generation, our approach employs a latent diffusion model to learn the joint distribution of cellular layouts, classification masks, and histology images. This model enables tailored data generation by conditioning on user-defined parameters such as cell types, quantities, and tissue types. Trained on the Conic H&E histopathology dataset and the Nissl-stained CytoDArk0 dataset, the model generates realistic and diverse labeled samples. Experimental results demonstrate improvements in cell instance segmentation and classification, particularly for underrepresented cell types like neutrophils in the Conic dataset. These findings underscore the potential of our approach to address data scarcity challenges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge