Turki Turki
A Noise-Filtering Approach for Cancer Drug Sensitivity Prediction
Dec 05, 2016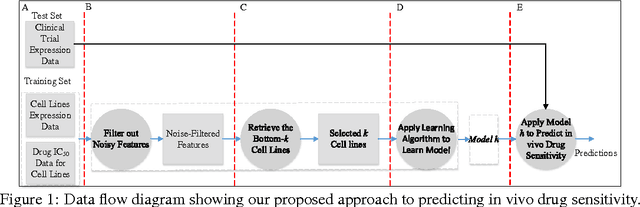
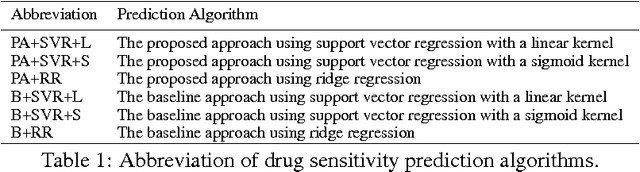
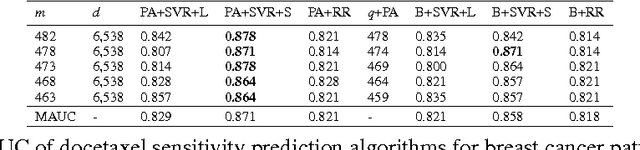
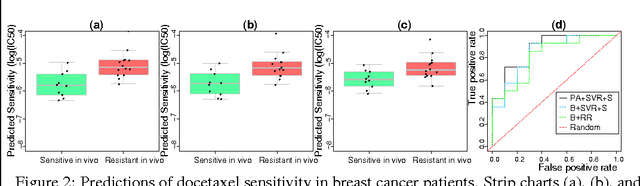
Abstract:Accurately predicting drug responses to cancer is an important problem hindering oncologists' efforts to find the most effective drugs to treat cancer, which is a core goal in precision medicine. The scientific community has focused on improving this prediction based on genomic, epigenomic, and proteomic datasets measured in human cancer cell lines. Real-world cancer cell lines contain noise, which degrades the performance of machine learning algorithms. This problem is rarely addressed in the existing approaches. In this paper, we present a noise-filtering approach that integrates techniques from numerical linear algebra and information retrieval targeted at filtering out noisy cancer cell lines. By filtering out noisy cancer cell lines, we can train machine learning algorithms on better quality cancer cell lines. We evaluate the performance of our approach and compare it with an existing approach using the Area Under the ROC Curve (AUC) on clinical trial data. The experimental results show that our proposed approach is stable and also yields the highest AUC at a statistically significant level.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge