Thayne Woycinck Kowalski
A Hybrid Ensemble Feature Selection Design for Candidate Biomarkers Discovery from Transcriptome Profiles
Jul 31, 2021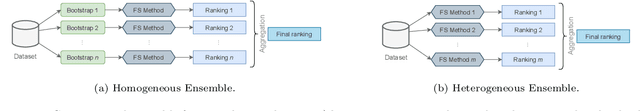

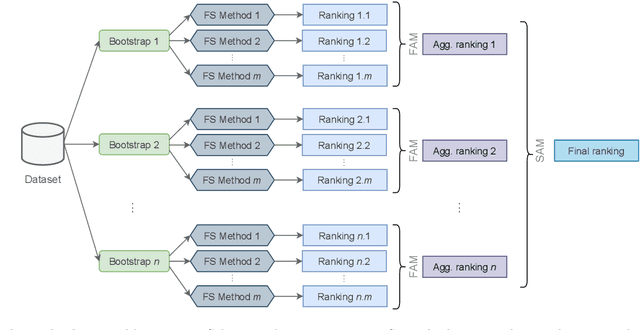
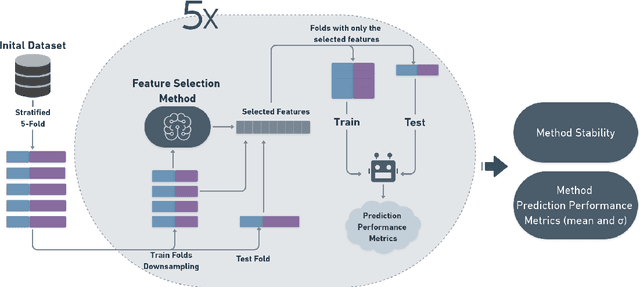
Abstract:The discovery of disease biomarkers from gene expression data has been greatly advanced by feature selection (FS) methods, especially using ensemble FS (EFS) strategies with perturbation at the data level (i.e., homogeneous, Hom-EFS) or method level (i.e., heterogeneous, Het-EFS). Here we proposed a Hybrid EFS (Hyb-EFS) design that explores both types of perturbation to improve the stability and the predictive power of candidate biomarkers. With this, Hyb-EFS aims to disrupt associations of good performance with a single dataset, single algorithm, or a specific combination of both, which is particularly interesting for better reproducibility of genomic biomarkers. We investigated the adequacy of our approach for microarray data related to four types of cancer, carrying out an extensive comparison with other ensemble and single FS approaches. Five FS methods were used in our experiments: Wx, Symmetrical Uncertainty (SU), Gain Ratio (GR), Characteristic Direction (GeoDE), and ReliefF. We observed that the Hyb-EFS and Het-EFS approaches attenuated the large performance variation observed for most single FS and Hom-EFS across distinct datasets. Also, the Hyb-EFS improved upon the stability of the Het-EFS within our domain. Comparing the Hyb-EFS and Het-EFS composed of the top-performing selectors (Wx, GR, and SU), our hybrid approach surpassed the equivalent heterogeneous design and the best Hom-EFS (Hom-Wx). Interestingly, the rankings produced by our Hyb-EFS reached greater biological plausibility, with a notably high enrichment for cancer-related genes and pathways. Thus, our experiments suggest the potential of the proposed Hybrid EFS design in discovering candidate biomarkers from microarray data. Finally, we provide an open-source framework to support similar analyses in other domains, both as a user-friendly application and a plain Python package.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge